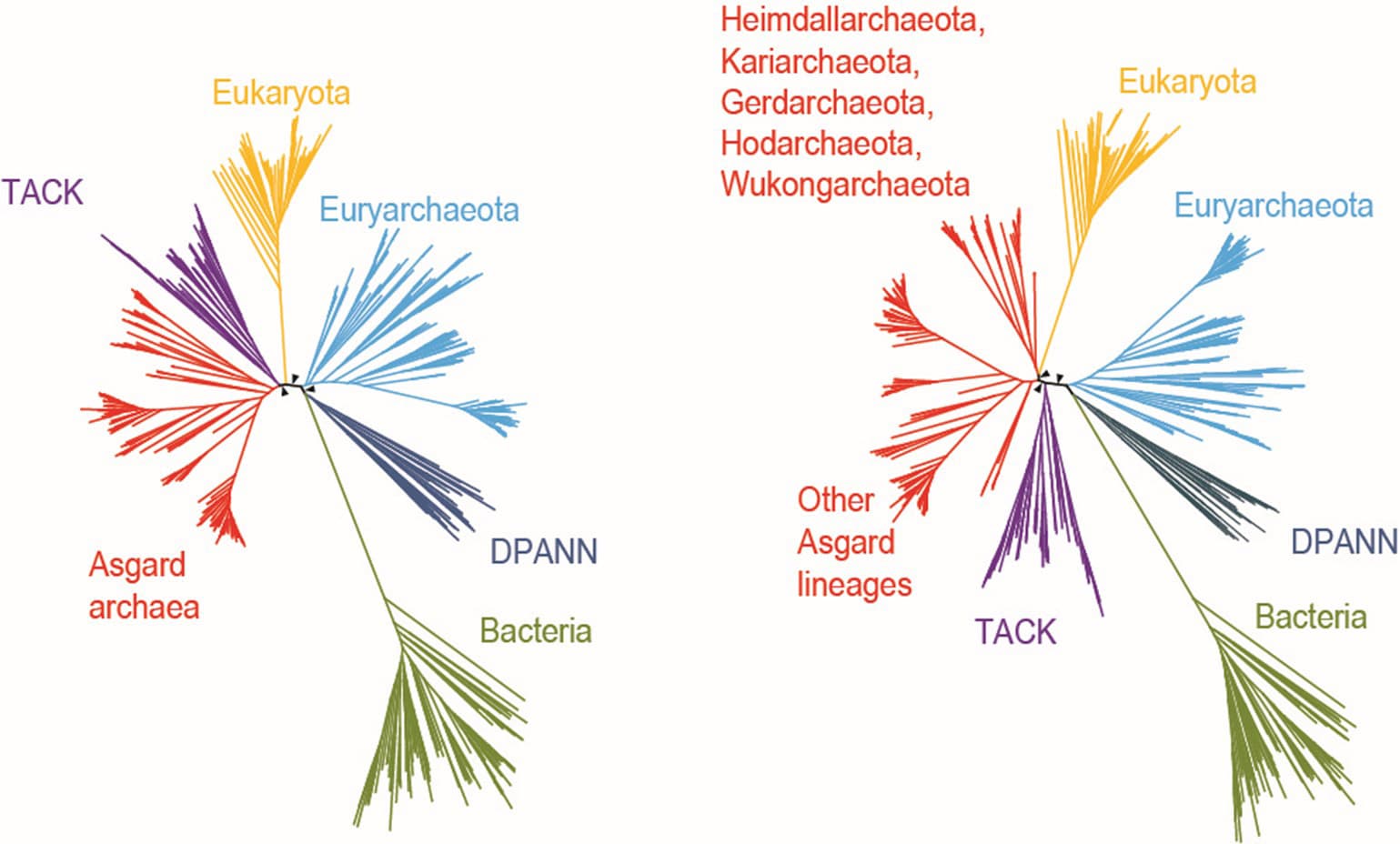
Phylogenetic tree of the Asgard archaea. Names in red are the novel phyla proposed. The cartoon of Wukongarchaeota is shown on the right. Browse Data from eLMSG (An eLibrary of Microbial Systematics and Genomics) via LMSG_G000000521.1-LMSG_G000000609.1
Prof. Meng Li’s group publishes a research article on Asgard archaea in Nature
Prof. Meng Li and his team from Institute for Advanced Study, Shenzhen University, have published an article entitled “Expanded diversity of Asgard archaea and their relationships with eukaryotes” in Nature on April 28th, 2021. In the study, they found six new Asgard phyla from various samples, including coastal sediments from Shenzhen Mangrove Natural Reserve and hadal seawater from the western Pacific Ocean. They constructed the first Asgard archaea cluster of orthologous gene (AsCOG) database. By analyzing the metabolic potentials of Asgard archaea and reconstructing the Tree of Life for Bacteria, Archaea and Eukaryotes, they elucidated the relationship between Asgard archaea and Eukaryotes in terms of the phylogeny, metabolisms and the functional evolution of Eukaryotic specific signature proteins, providing new insights into the unresolved basic scientific question - Eukaryogenesis.
This research involved the collaboration of Shenzhen University, National Institutes of Health (USA), Third Institute of Oceanography of Ministry of Natural Resources and the Biogas Institute of Ministry of Agriculture. Prof Meng Li from Institute for Advanced Study at Shenzhen University and Prof. Eugene Koonin from National Institutes of Health (USA) are the corresponding authors. Associate research fellow Dr. Yang Liu (Shenzhen University), Dr. Kira Makarova (National Institutes of Health; USA), and Mr. Wen-Cong Huang (Master student at Shenzhen University) are the co-first authors.
Asgard is the fourth superphylum of the Archaea domain. Based on the phylogenetic analysis of genomic data, some scholars proposed that Eukaryotes originate from Asgard archaea. In 2019, a Japanese research group firstly reported an enrichment culture of Lokiarchaeota in the laboratory. The Science journal listed the study of Asgard archaea and eukaryogenesis as the top ten significant scientific breakthroughs of the year 2019. Therefore, studies on Asgard archaea and Eukaryogenesis have become the frontier and hotspot of biological sciences.
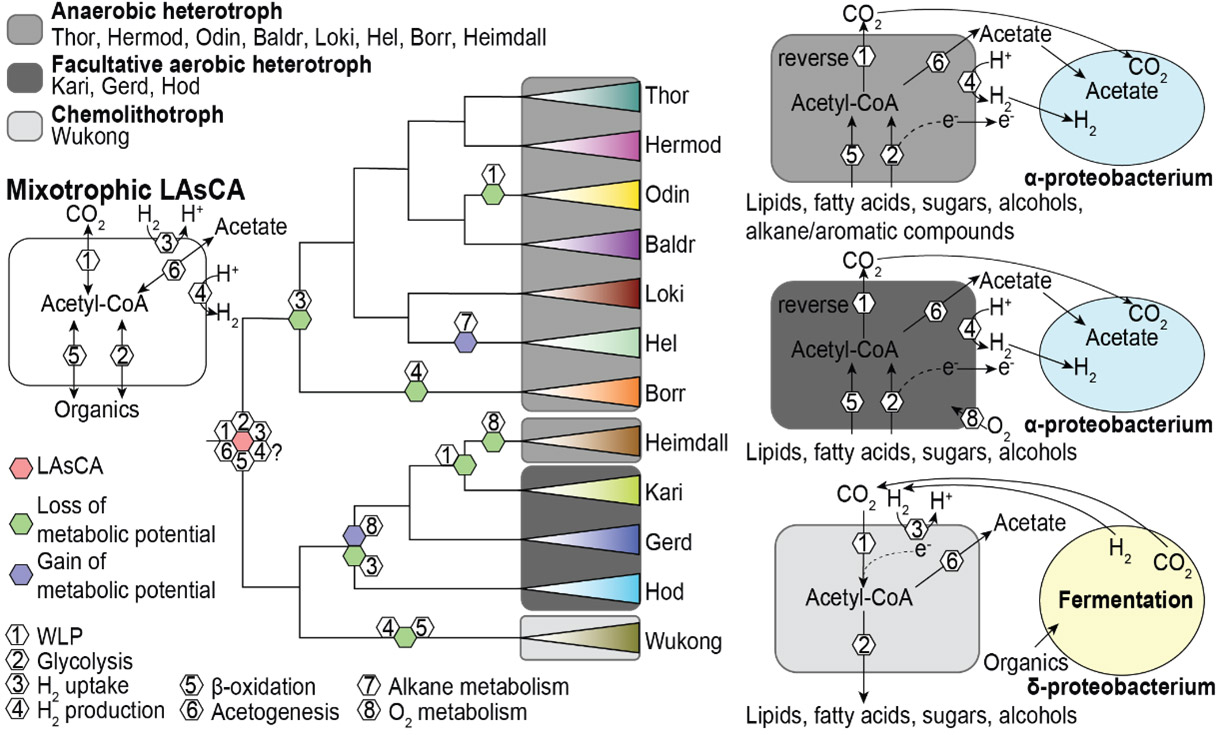
The Asgard archaea initially included five phyla, including Loki-, Thor-, Odin-, Heimdall- and Helarchaeota. Prof Meng Li’s group identified the sixth Asgard archaea phylum named Gerdarchaeota in mangrove sediments in early 2020. Collaborating with Eugene’s group, they reported 75 novel Asgard MAGs, which substantially expand the phylogenetic diversity of Asgard, and proposed six additional phyla. Five phyla are named after the Norse deities, i.e., Hermod-, Baldr-, Borr-, Kari-, Hod- and a deep branch Wukongarchaeota is named after the Chinese mythological character “The Monkey King – Sun Wukong”. The metabolic reconstruction suggests that Wukongarchaeota is an obligate hydrogenotrophic acetogen, which is dramatically different from other known Asgard archaea. This is why it was named after a Chinese mythological character, the departure from the Norse Pantheon for Asgard archaea.
This research is supported by National Natural Science Foundation of China, the Innovation Team Project of Universities in Guangdong Province and the Shenzhen Science and Technology Program.
Find the article at https://www.nature.com/articles/s41586-021-03494-3 and the story behind the paper at https://natureecoevocommunity.nature.com/posts/exploring-asgard-archaea-and-their-relationships-with-eukaryotes .