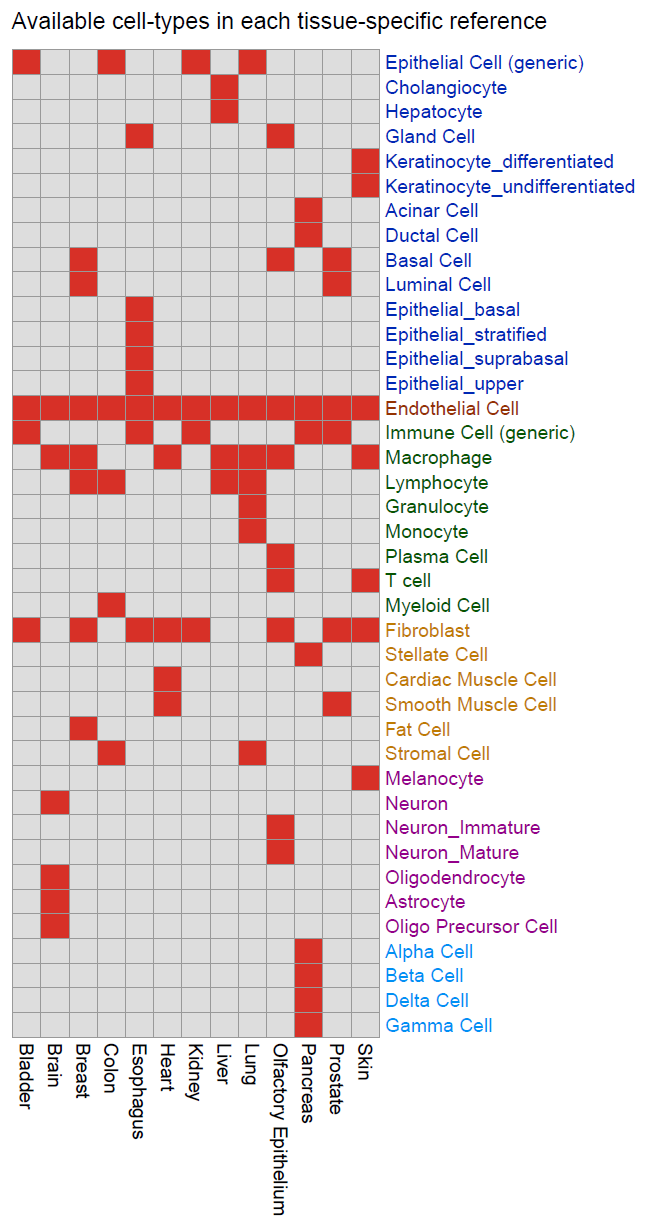
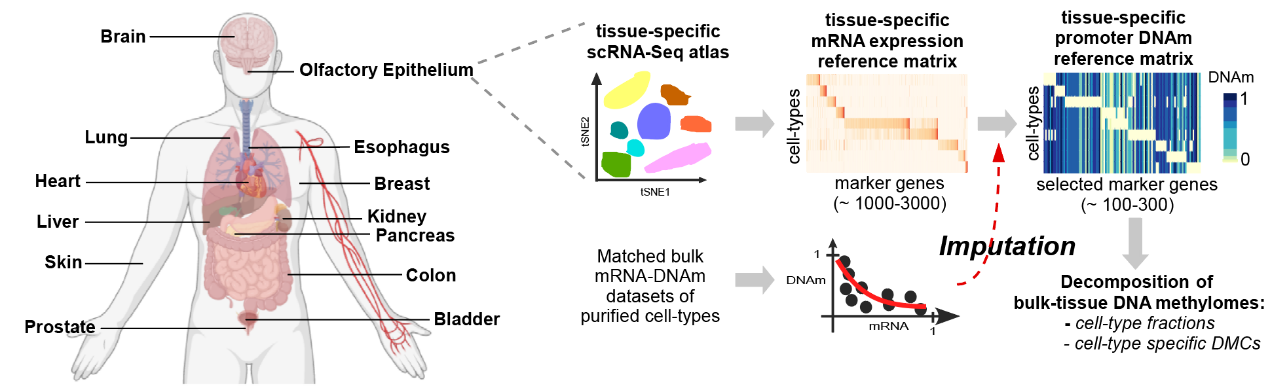
We hope this database will be a useful resource for:
- Estimation of all main cell-type fractions in a given tissue
- Detection of differentially methylated cytosines independent of cell-type composition
- Detection of cell-type specific differential DNAm
Suggested tools to run cell-type deconvolution:
- EpiSCORE R package: https://github.com/aet21/EpiSCORE
- EpiDISH web server: https://www.biosino.org/EpiDISH/
Note: For both tools, the DNAm-atlas reference matrices are pre-installed in the package/web server and can be applied directly.