How to use our database
To achieve a better performance, we recommend you to use a latest browser.
Nevertheless, a browser with higher version than Chrome 15, Internet Explorer 10, Firefox 8, Opera 12, Safari 8 will work fine.
CamelDB database is designed to provide comprehensive resources for basic science and applied medicine
research based on camelid genome, which may offer considerable value in various scientific
and applied fields such as evolutionary biology, metabolic adaptation to extreme environment,
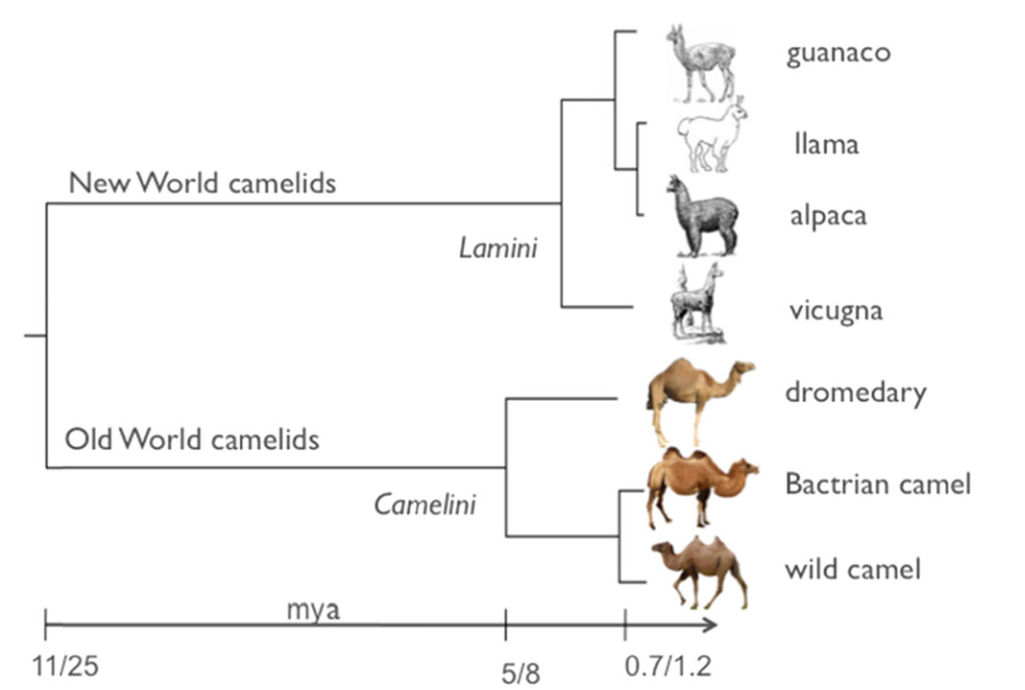
agricultural breeding and antibody engineering. You can overview the evolutionary tree of
seven species in Camlid which collected in our database.
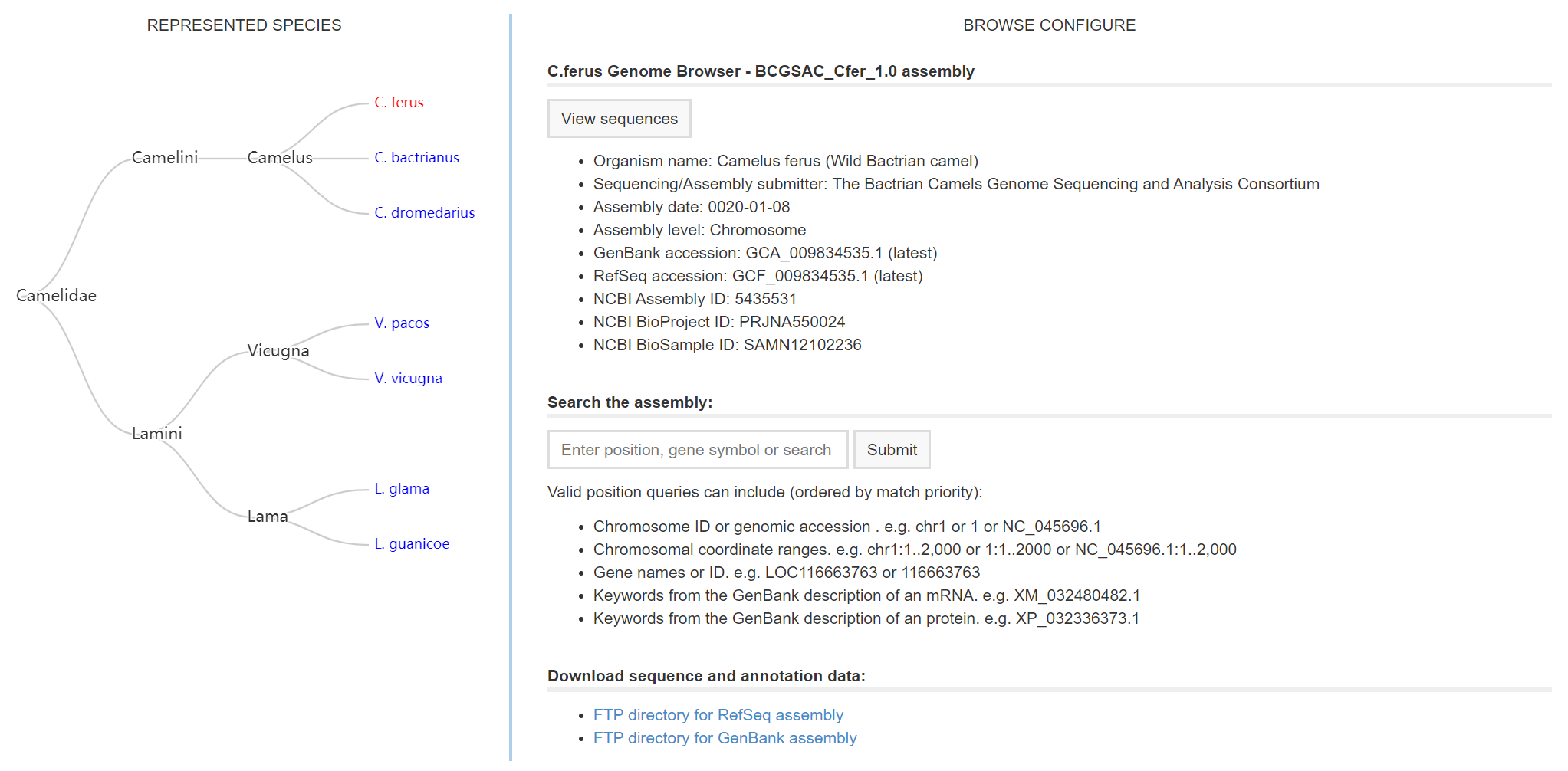
Genome assemblies browse and report page
In the page left, you can choose one of seven species to browse the genome assembly and to search feature type in our database.
In the page right, you can click the View sequences to browse all the chromosome or scaffold in chosen species.
Also, you can enter a feature item in the Search box to search corresponding sequence information.
If multiple query records are returned, results will be returned in a table in which each row represents a chromome or scaffold including some summary information.
Click links on result page will lead you to the corresponding JBrowse report page.
Otherwise, a corresponding JBrowse report page will be display.
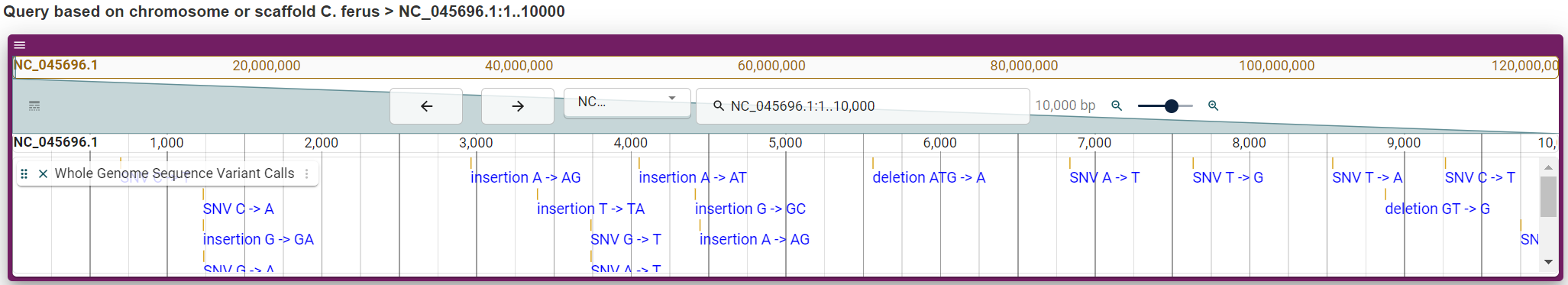
Report page
You can use the query frames to understand the local synteny in gene level among C. ferus, C. dromedarius and V. pacos.
Just select an frame you concerned in the search
form and choose corresponding items, then press enter or click the submit button,
the results will be displayed in a few seconds.
Report page for each query frame
- SynCircos
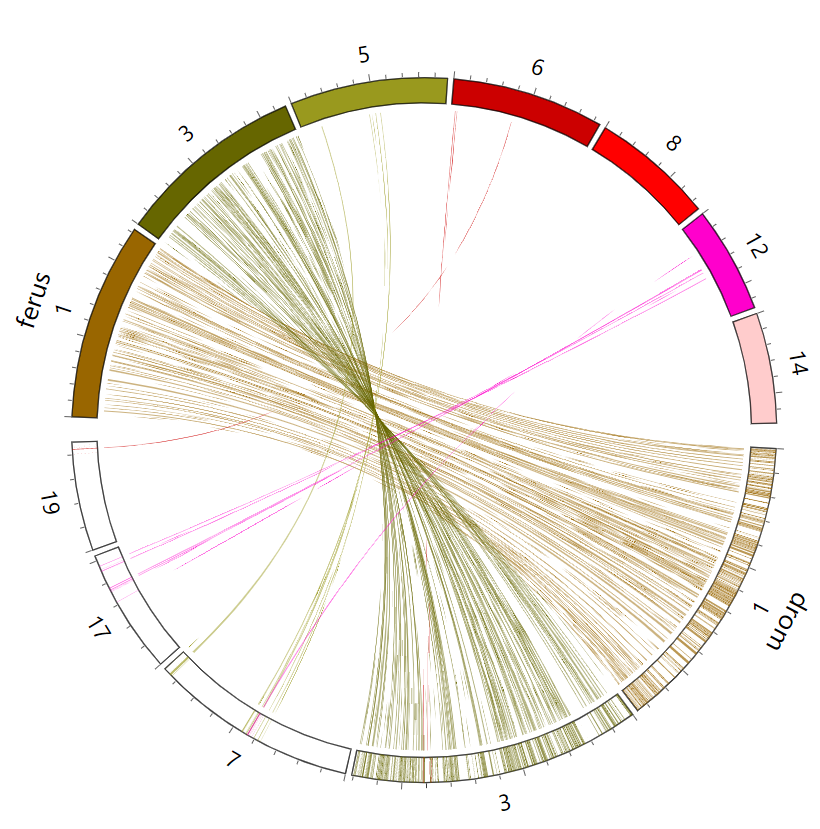
- SynBrowser
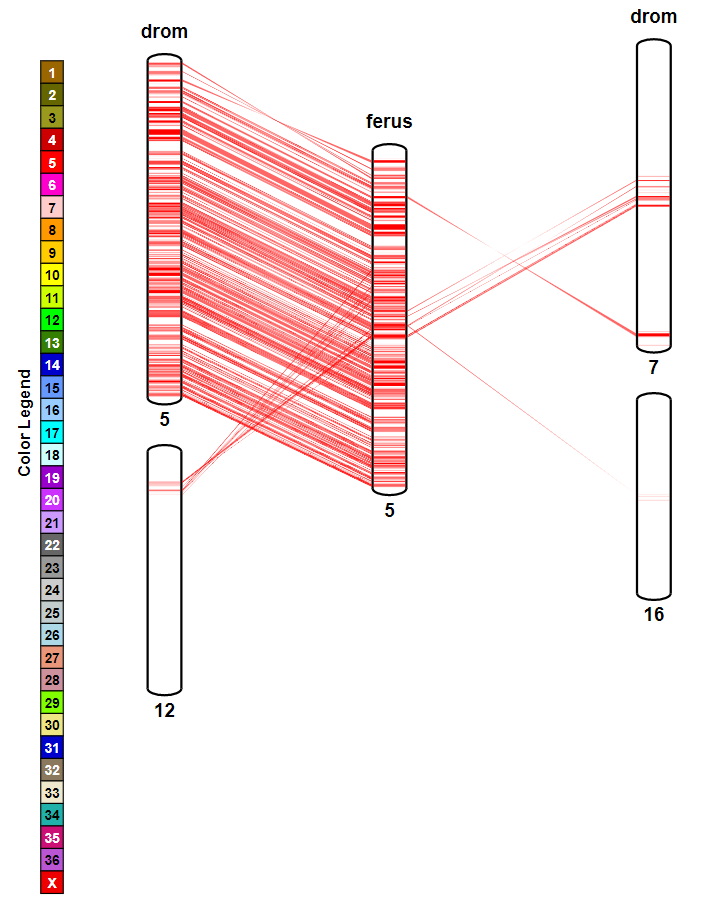
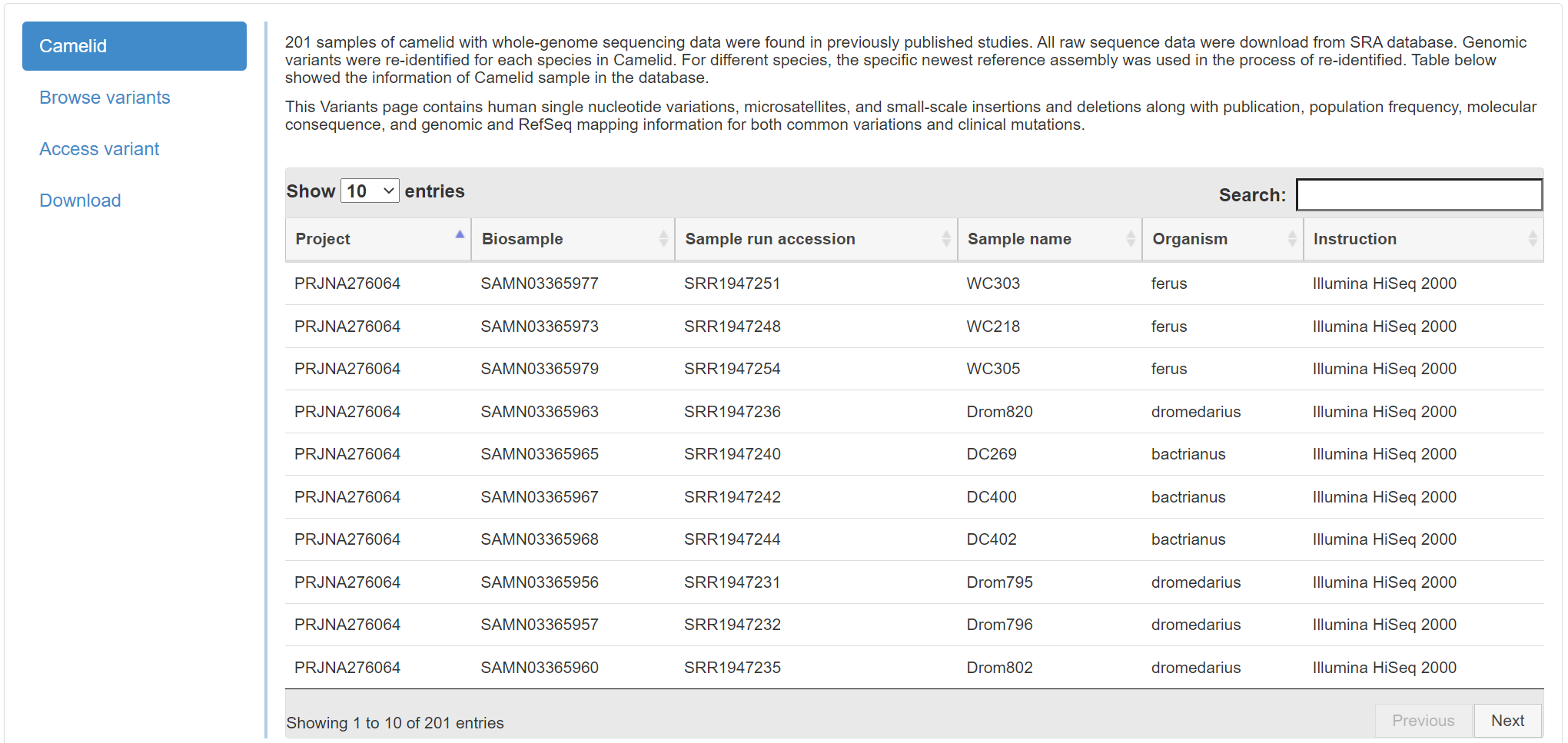
There are four frames in this page, you can use Browse variants, Access variant
to understand the genetic variants in out database.
Report page for each query frame
- Browse variants
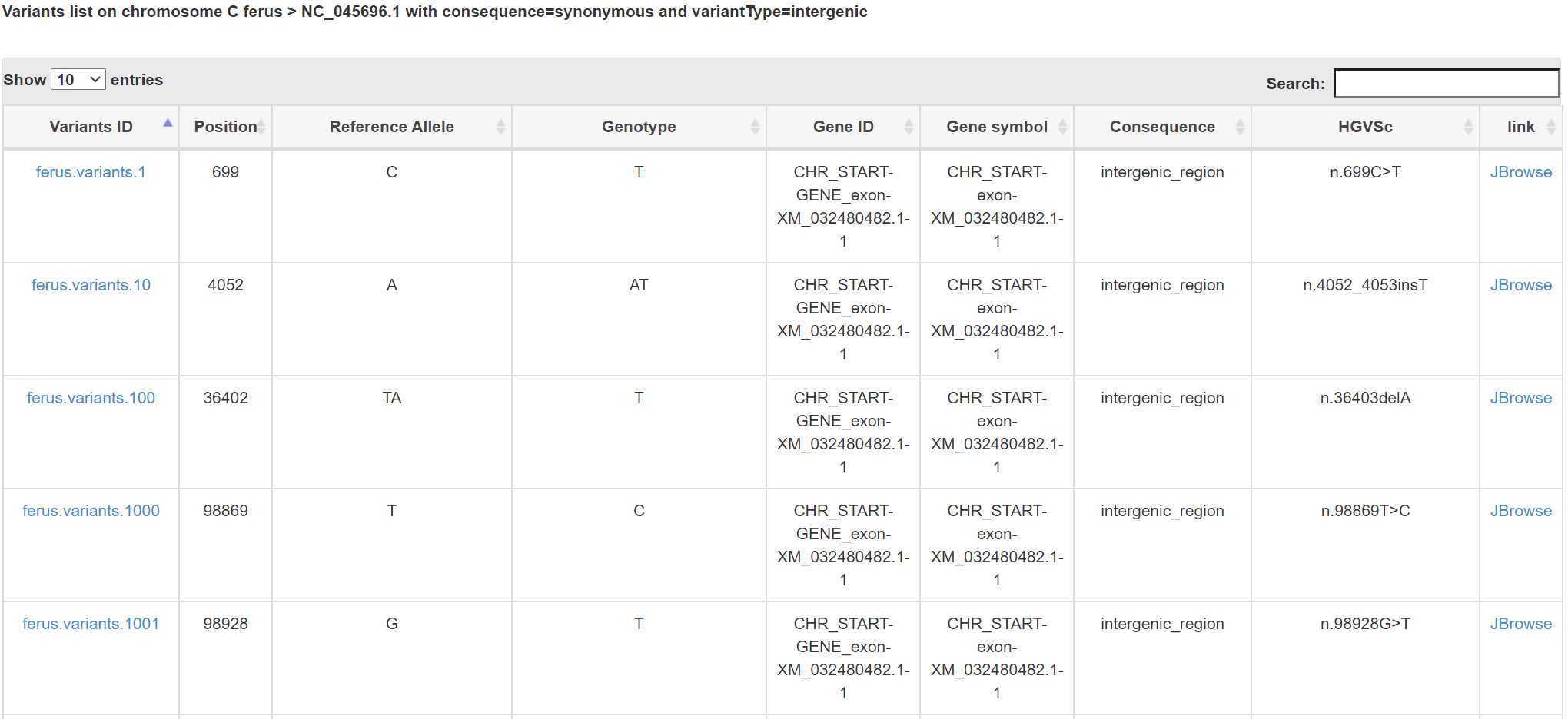
You can choose the combination of four conditions: species, Consequence, Variant type Chromosome or scaffold to browse genetic variants in our database. Click links on that page will lead you to the corresponding report page.
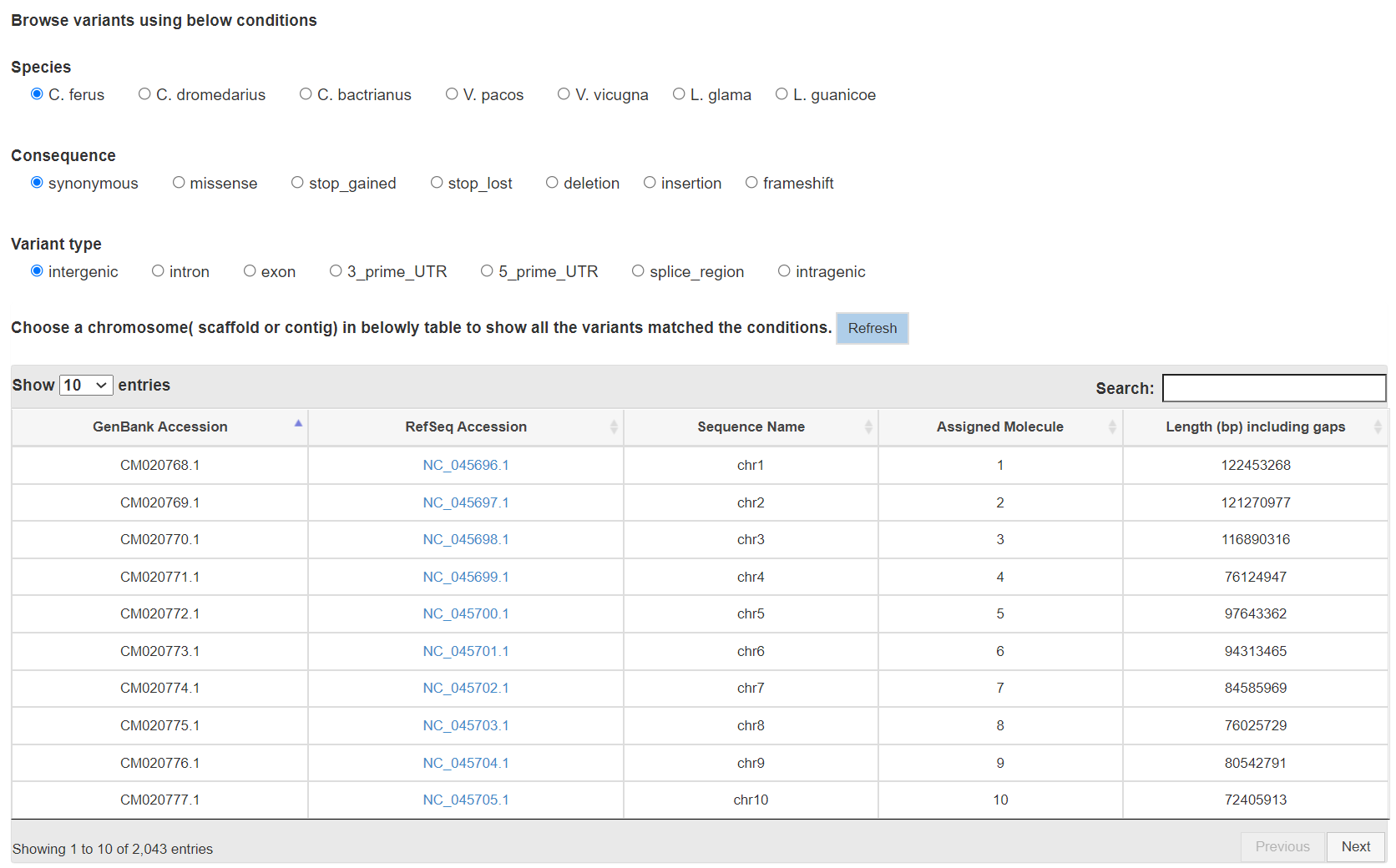
Results will be returned in a table in which each row represents a genetic variant including some detaily information.
- Search variant
You can choose one of four feture types: Variant ID, Gene symbol, Protein ID mRNA ID to search corresponding genetic variants in our database. Click links on that page will lead you to the corresponding report page.
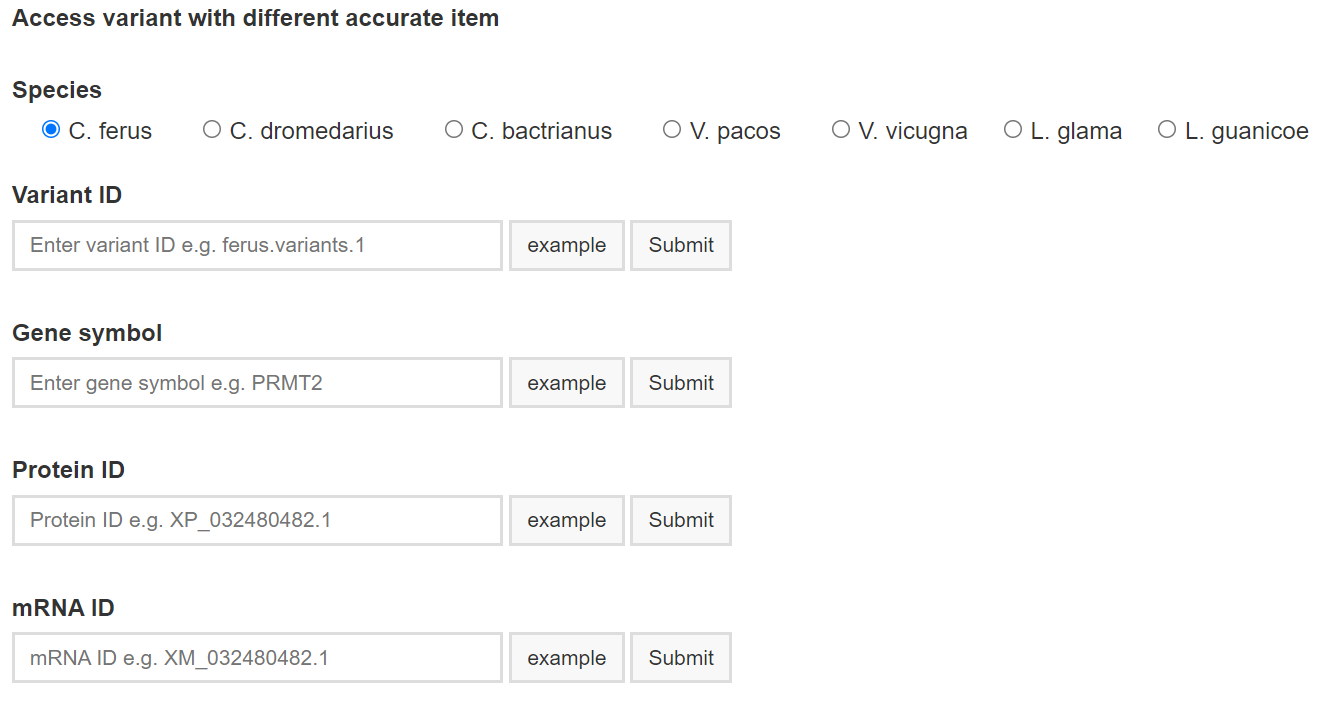
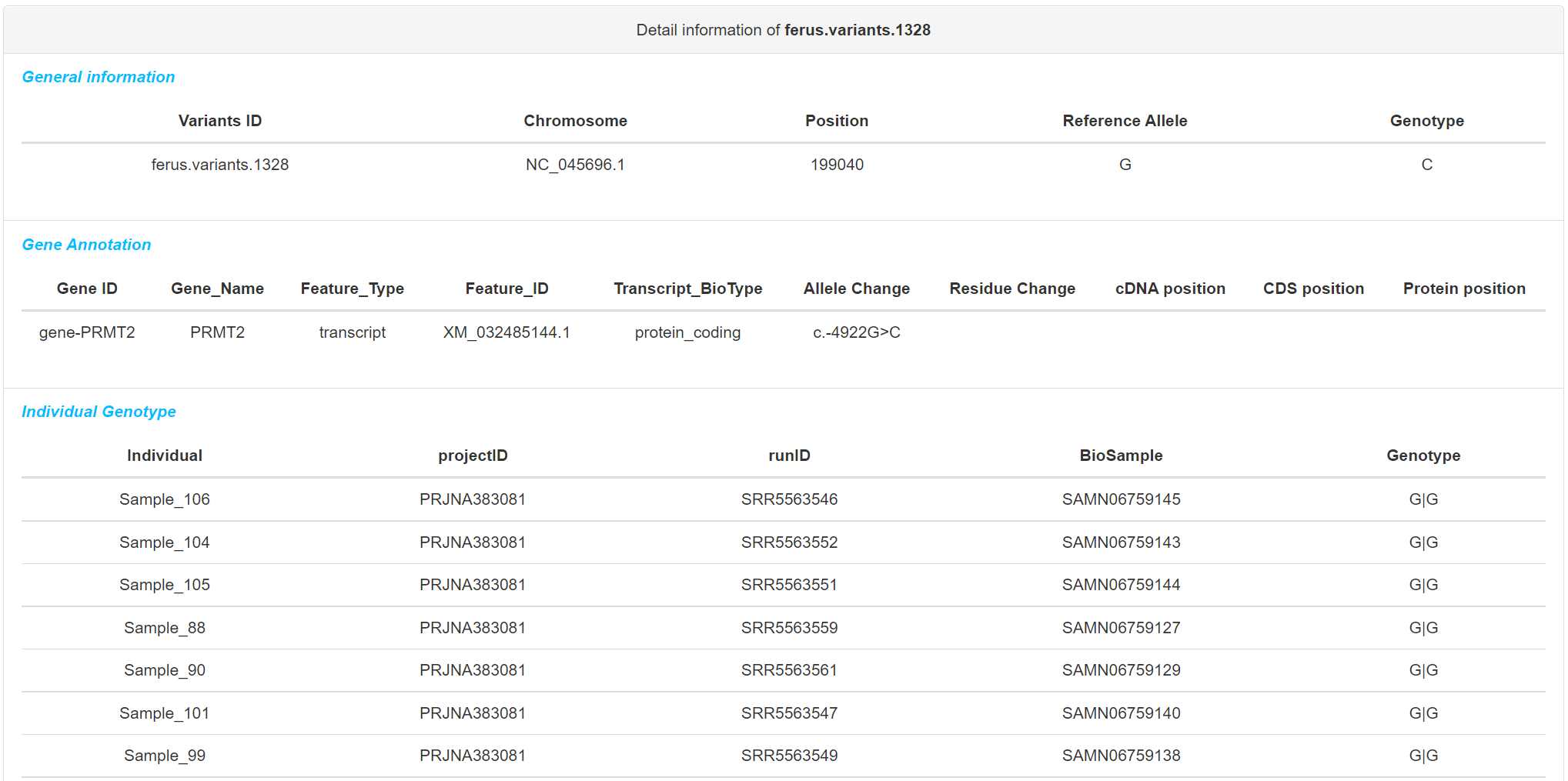
If multiple genetic variants are returned, results will be returned in a table in which each row represents a genetic variant including some detaily information. Otherwise, detail report of genetic variant will be display.
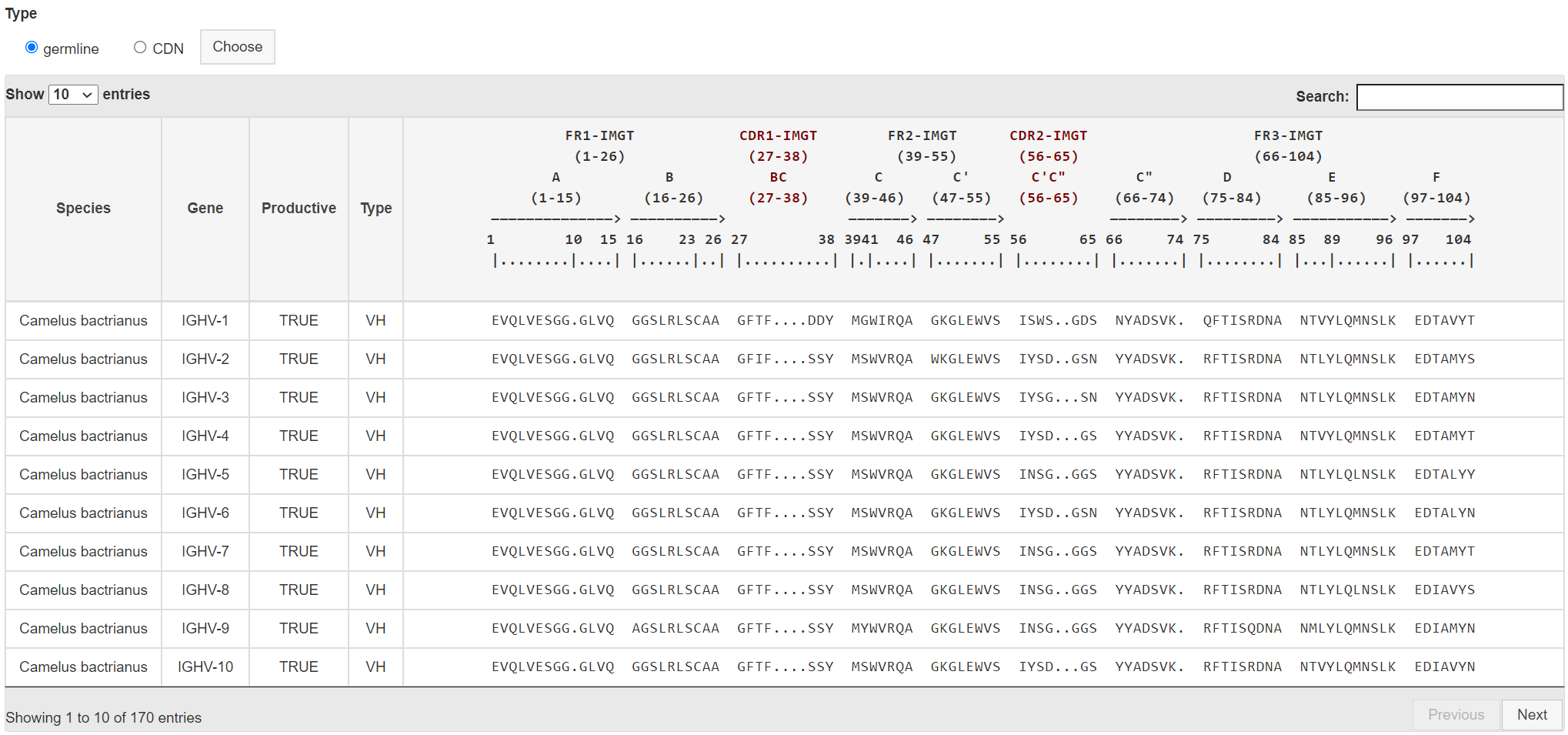
You can overview all antibody gene in this page. You can choose gDNA, cDNA
to display different antibody gene.
The table gives the detaily information of each antibody gene.
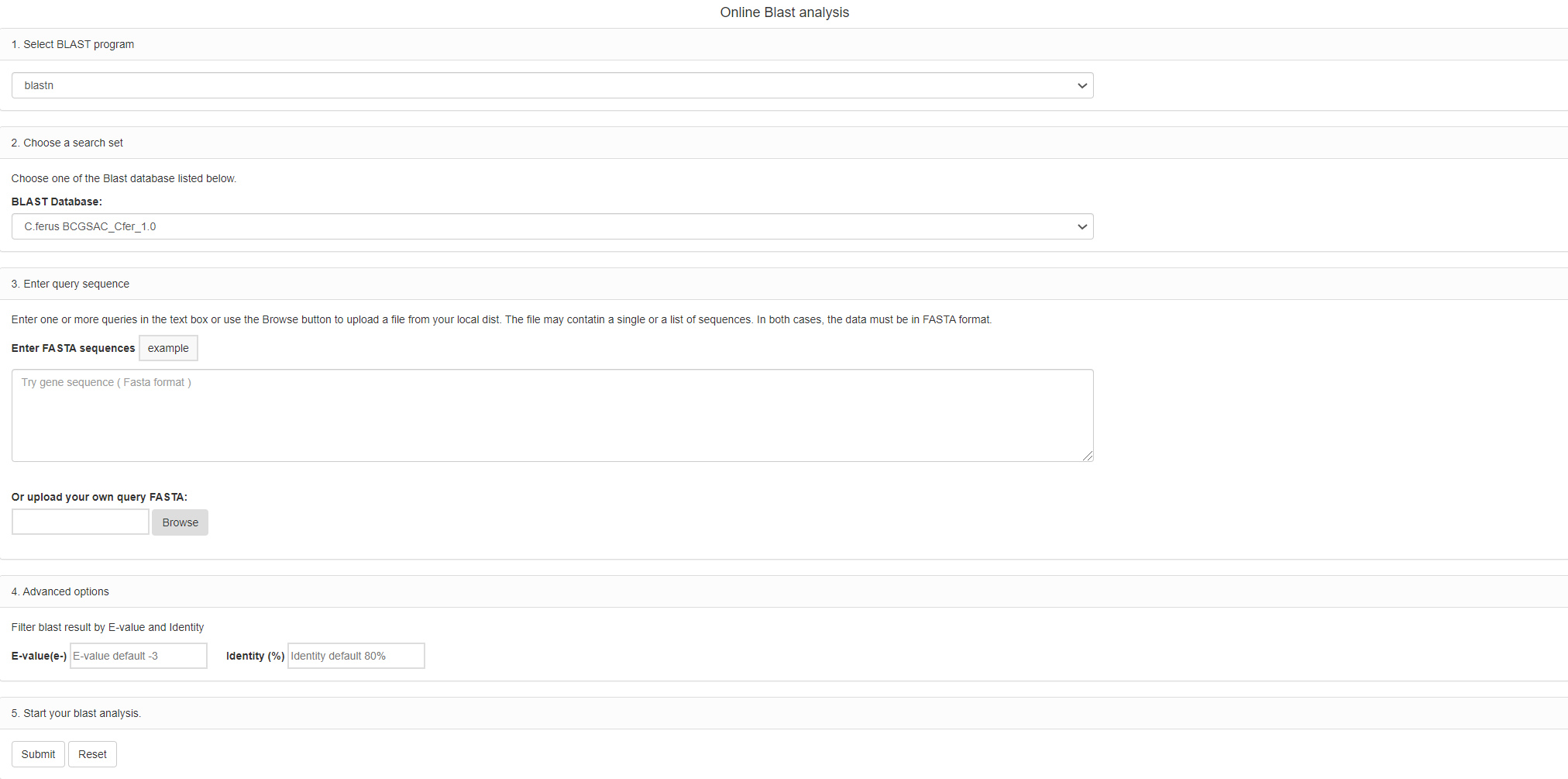
For a better understanding genomic information of species in camelid, we provide two conventional data analysis tools: BLAST and Imputation,
The first tool and report page: BLAST
You can intuitively use blast program between your sequence and reference genome sequence in species of camlid.
Report page
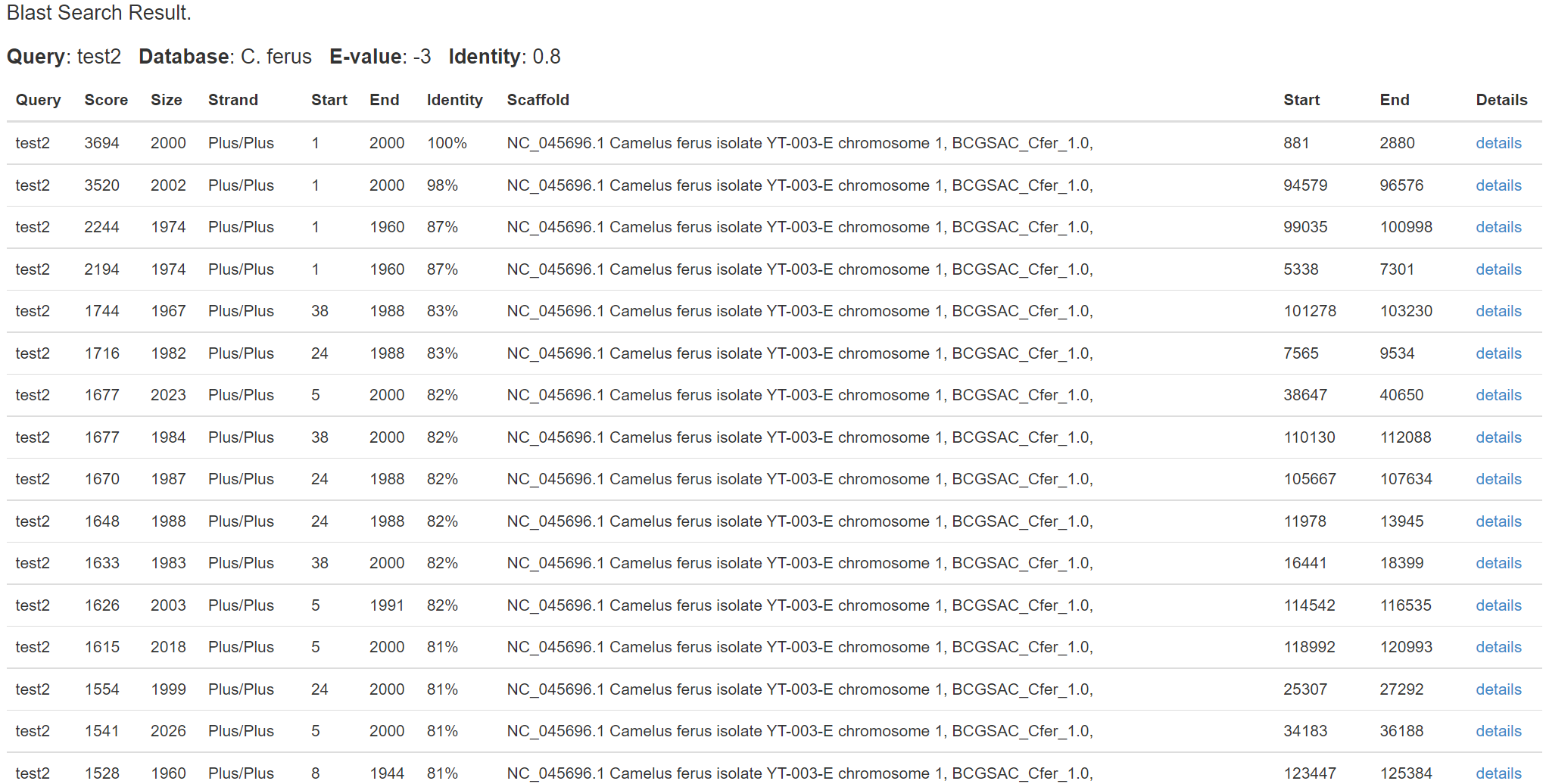
Attention: you can click the details to show the alignment.

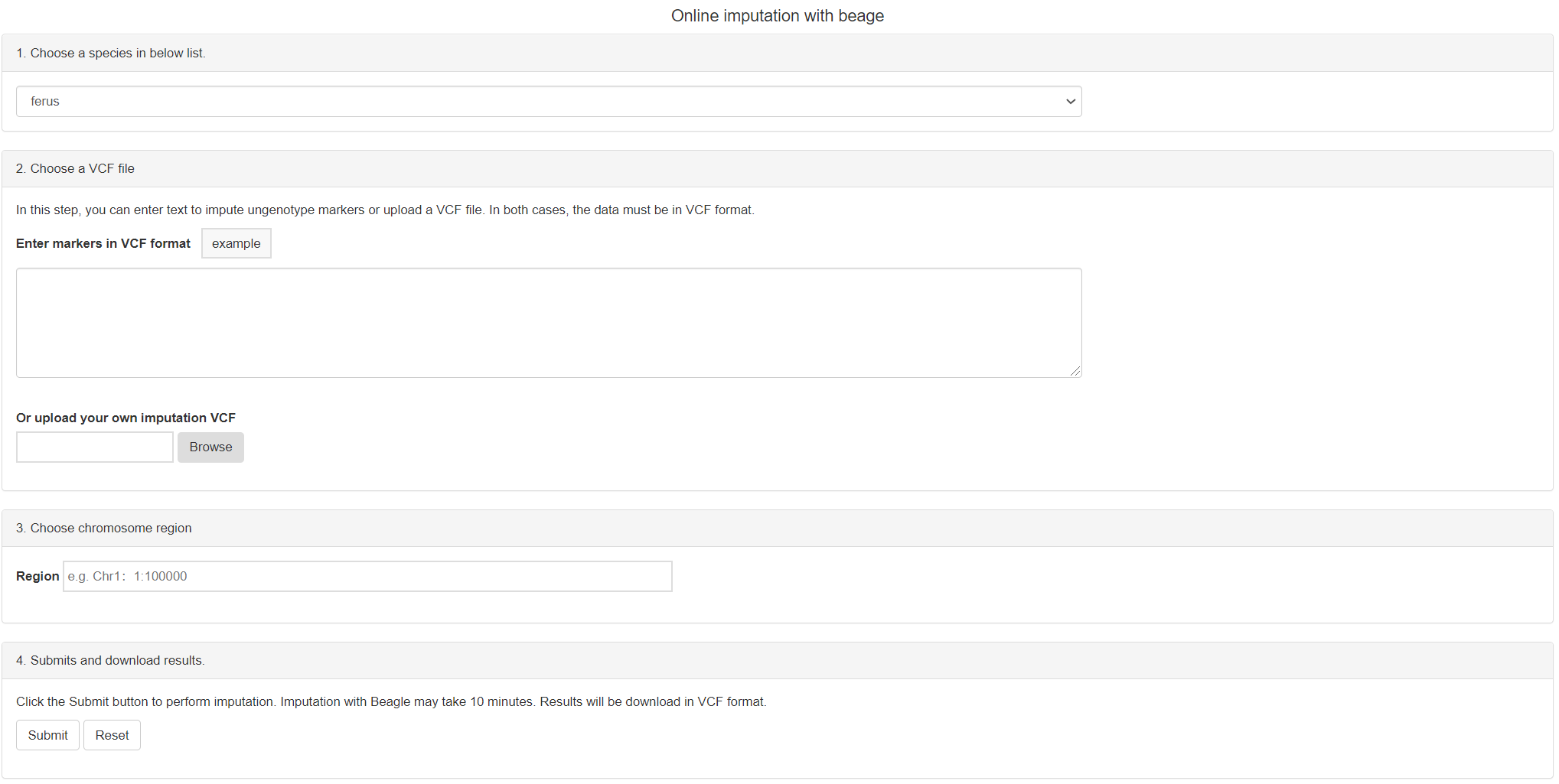
The second tool and report page: Imputation
You can use Beagle program for genotype imputation to facilitate GWAS research on camelid species. The result will be download as vcf file.