Users can search by drug, disease, gene, variant, phenotype or relation to get an interested knowledge list.
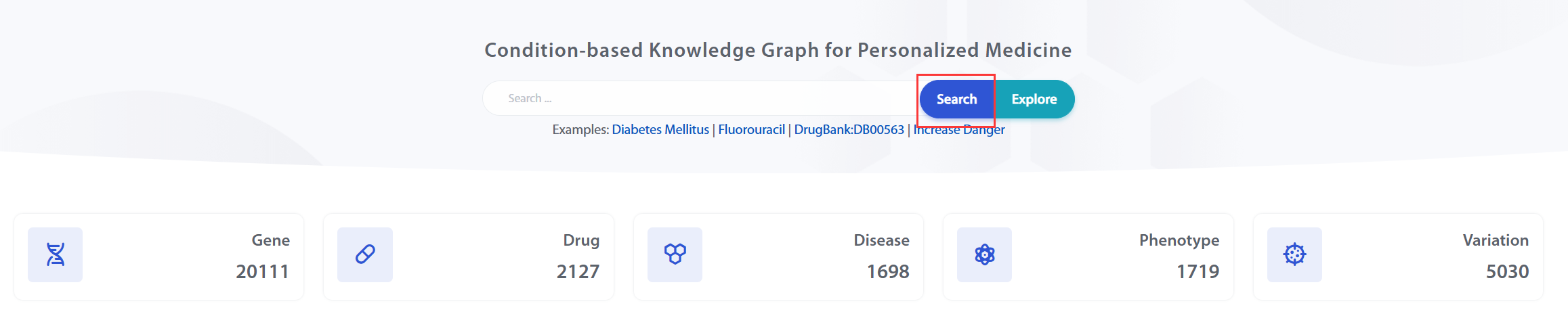
The search terms have an auto-complete feature, which uses standardized names and aliases for auto-completion. The final display will be in the format: [Standard Name [Type] (Typed Alias)].
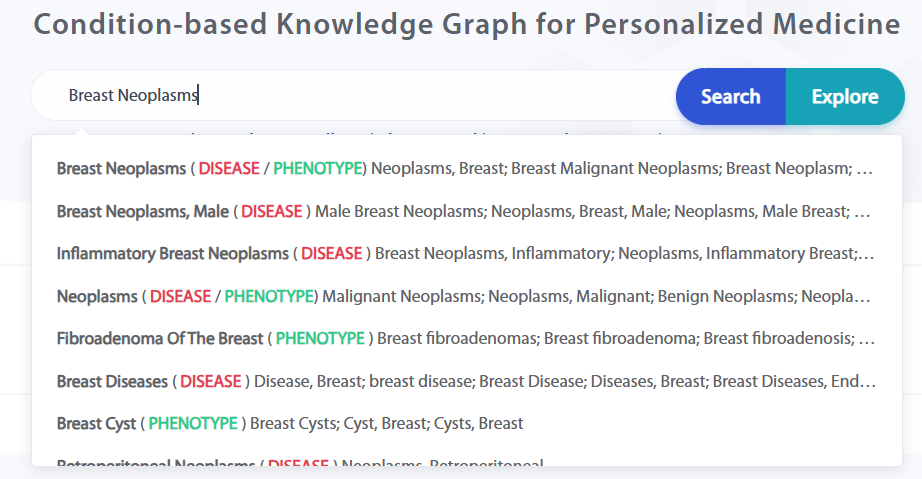
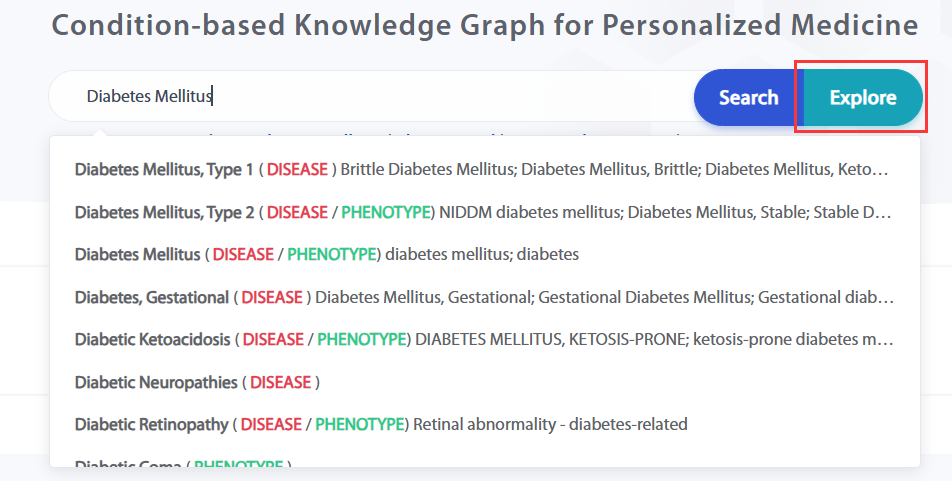
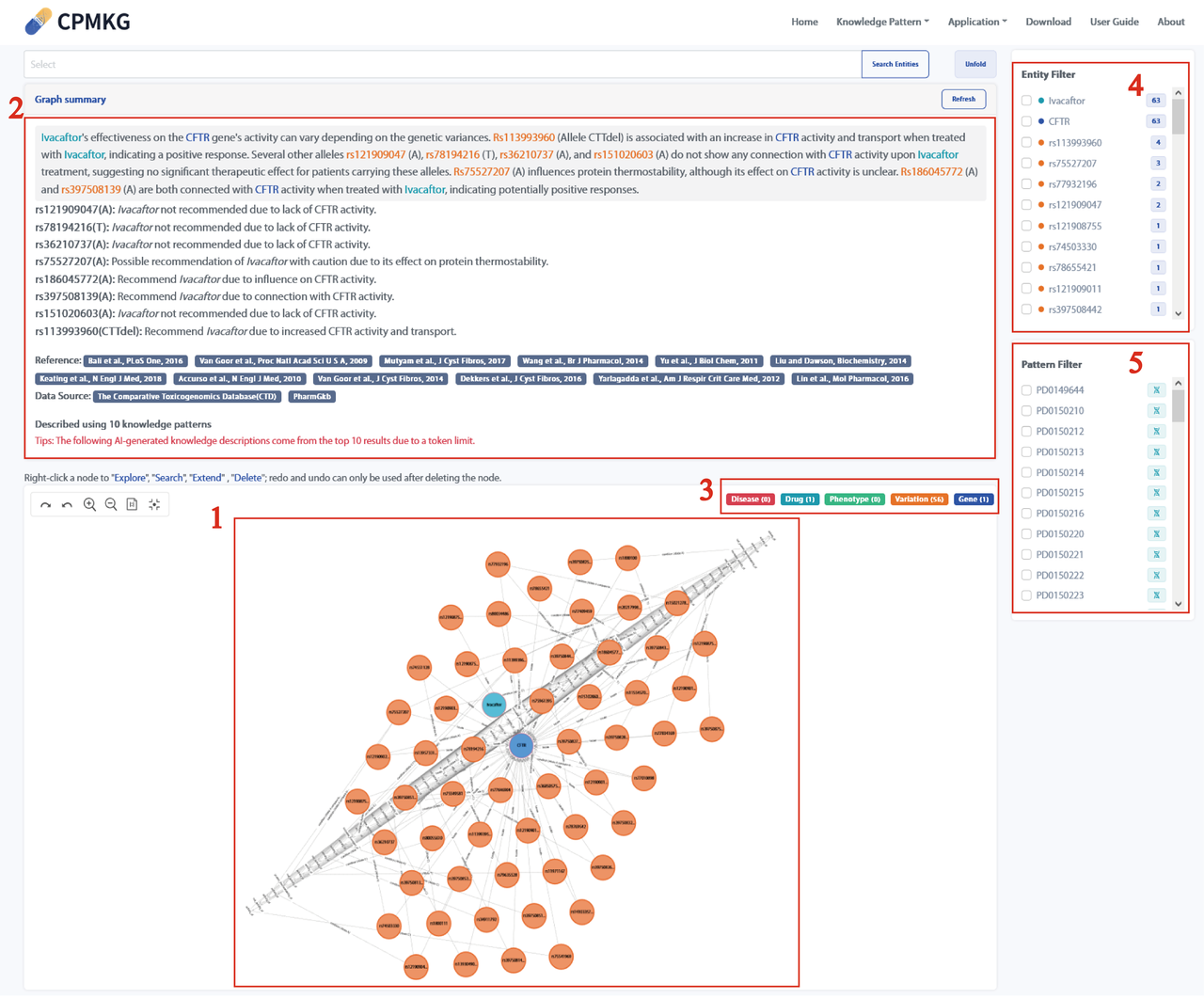
Users can search for knowledge related to this entity and present it in a graph using the "Explore" function.
The webpage content after clicking 'Explore' includes:
Refer to the webpage below for details.
Note: Due to the input token limit of the underlying LLM, subgraph descriptions are limited to the top 10 knowledge items in the search results. To clarify, we have added the following tip on our webpage: " The following AI-generated knowledge descriptions are based on the top 10 results due to the token limit."

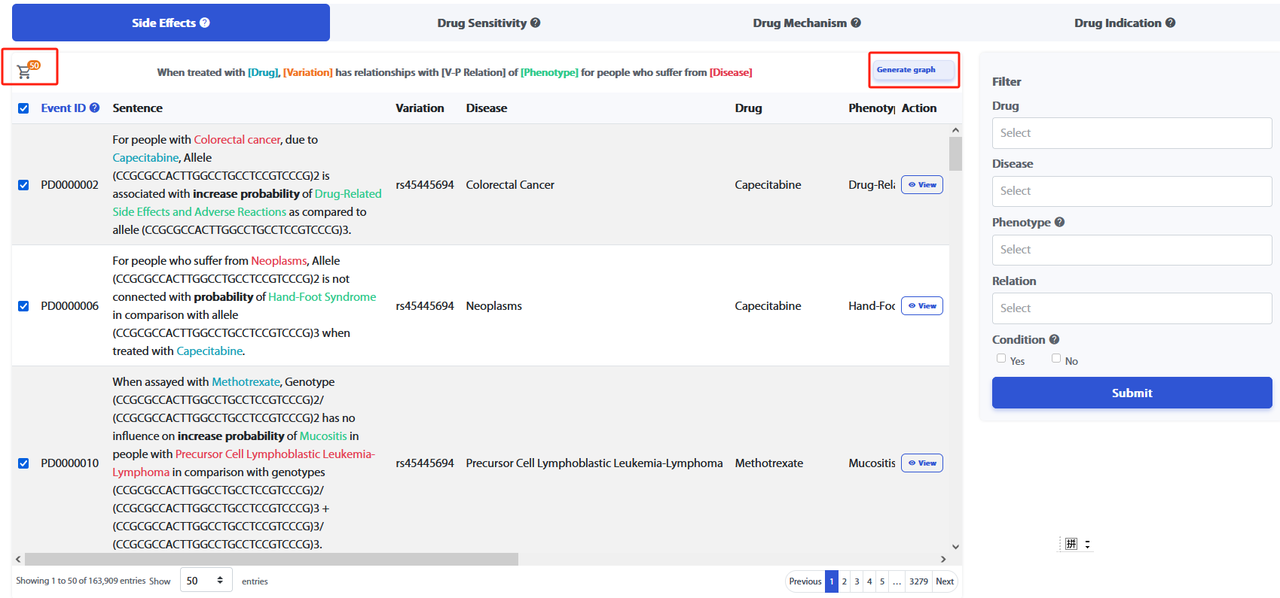
Users can acquire knowledge by conducting searches on the Home page or by clicking on the pattern name in the Navigation bar. The knowledge pattern list displays categories such as Side Effects, Drug Sensitivity, Drug Mechanism, and Drug Indication. By default, without applying any filters, data from all knowledge patterns will be shown. Users can switch between different lists by selecting various knowledge patterns. Refer to the figure below for more details.
On the right side, the "Filter" allows filtering of knowledge fitting the four knowledge patterns through Drug, Disease, Phenotype, Relation, and Condition. The Condition filter is selected by a "Yes/No" checkbox, with "Yes" as the default. Clicking "Yes" will bring up the Variation filter. Other entity filters are applied by users entering the names of entities they are interested in to further refine the knowledge of interest. Clicking "Submit" will display the corresponding filtered results.
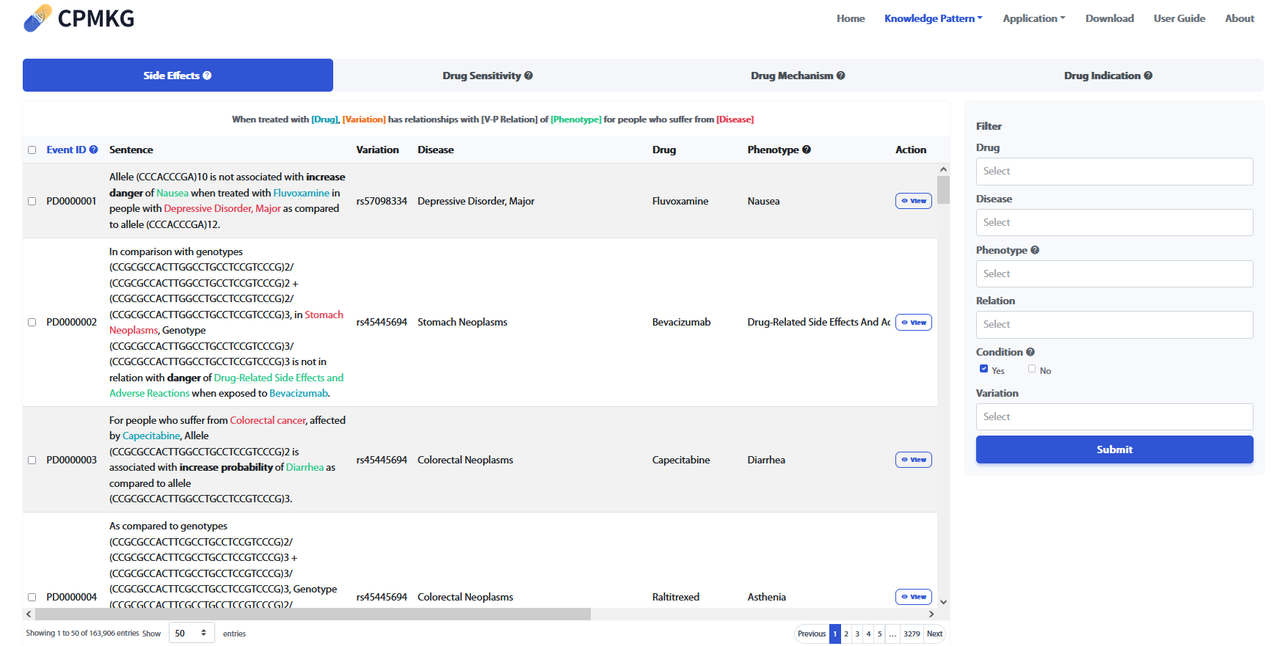
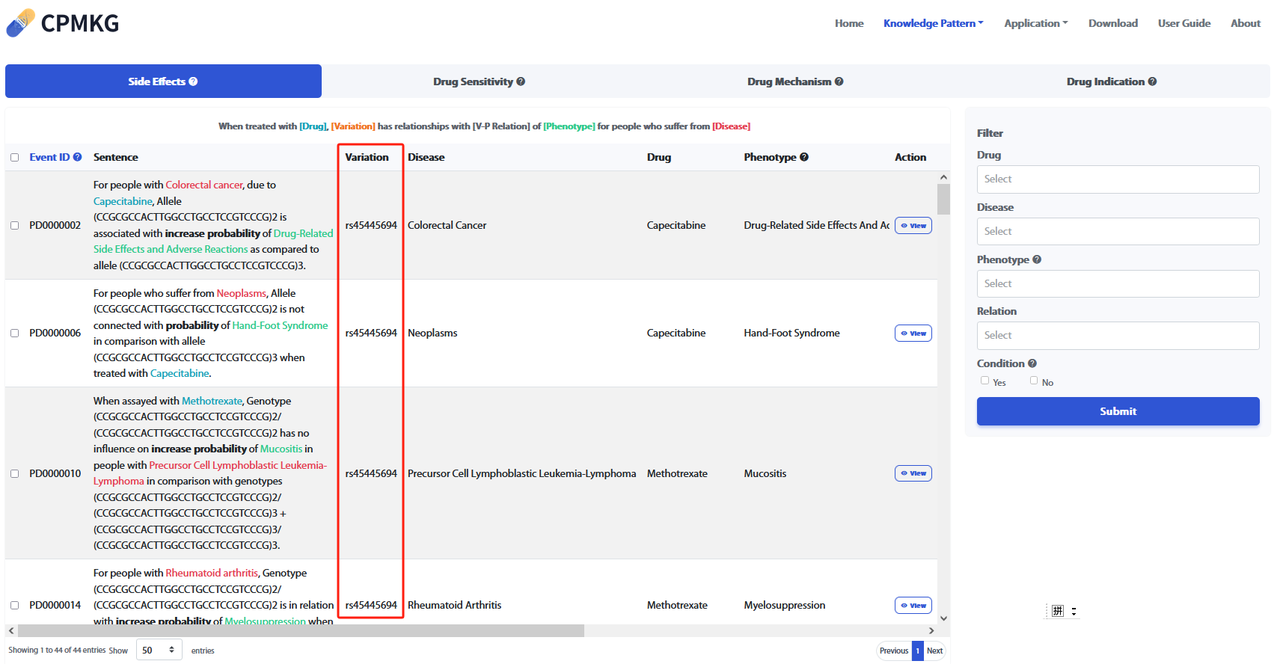
By clicking "View" under the "Action" column in the "Knowledge Pattern" list, you can view the details page of each piece of knowledge.

The details page displays comprehensive information about the knowledge, including:
Refer to the webpage below for more details.

By selecting the "Event ID" column in the "Knowledge Pattern" list and clicking "Generate Graph", users can display multiple pieces of knowledge. This feature supports the selection of Event IDs across different knowledge patterns.
The display of the webpage after clicking "Generate Graph" includes:
Refer to the webpage below for details.
Note: Due to the input token limit of the underlying LLM, subgraph descriptions are limited to the top 10 knowledge items in the search results. To clarify, we have added the following tip on our webpage: " The following AI-generated knowledge descriptions are based on the top 10 results due to the token limit."
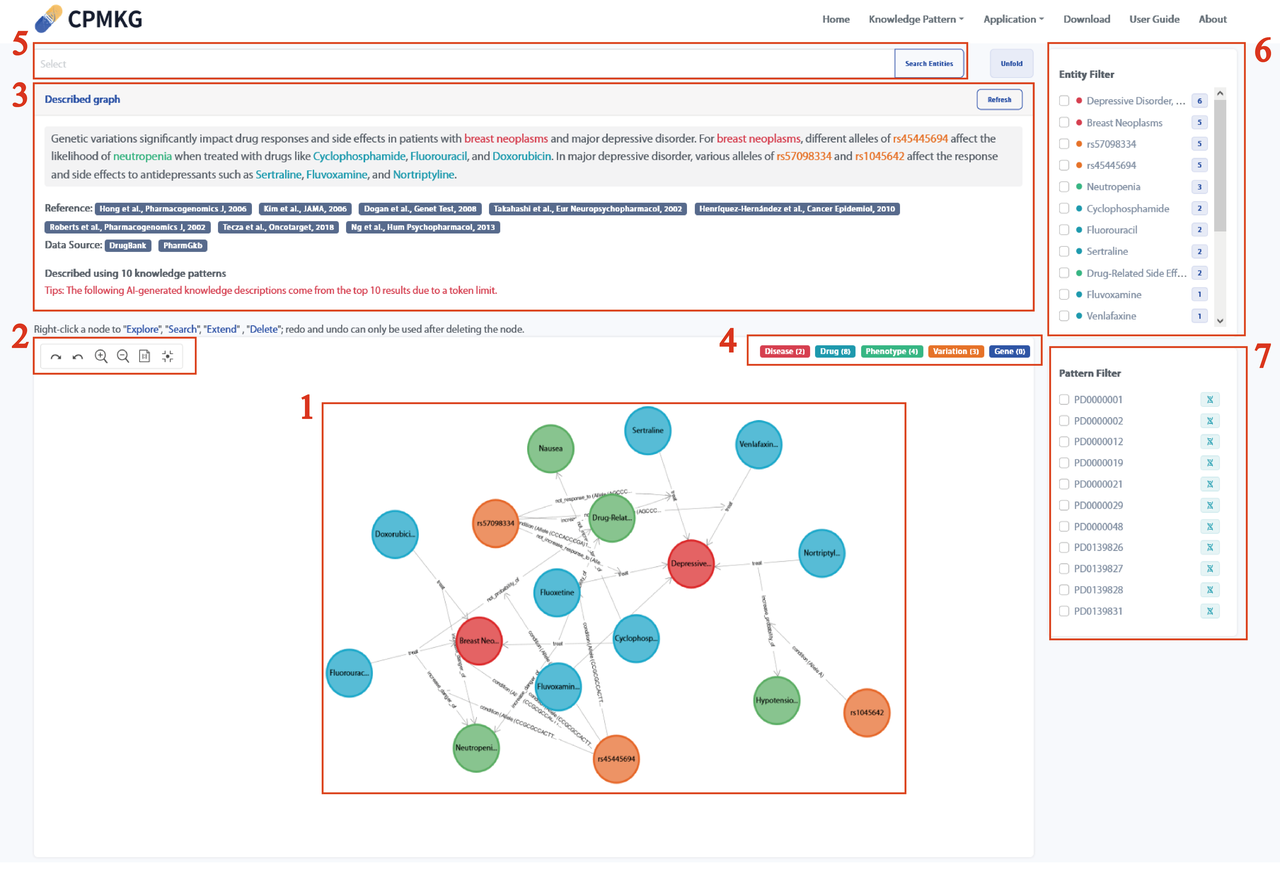
Hovering over a node in the knowledge graph displays detailed information about the node, including its category, ID, name, aliases, and description. Hovering over a relationship edge will show the relation, Event ID, and data source, with the data source being clickable. Refer to the webpage below for more details.

Left-clicking on a node in the knowledge graph will display the following options: Explore (show a subgraph centered on this node), Search (search for this node, which will redirect to the "Knowledge Pattern" page), Extend (expand to include other nodes connected to this node), and Delete (remove this node along with its related connections and other nodes).
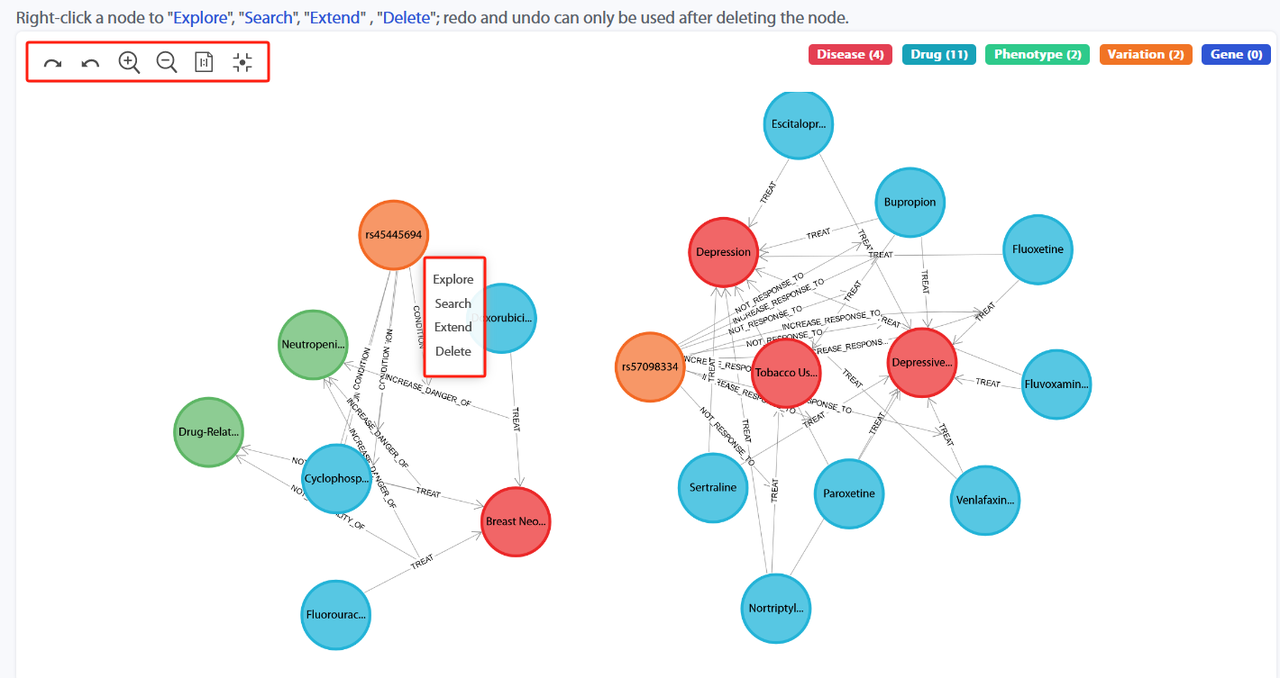
For example, when clicking "Explore" on "rs45445694", which will show the result on the webpage below.

Using "rs45445694" as an example, click "Search" and the results will be shown as illustrated below.
Using 'rs45445694' as an example, click 'Extend' and the result is shown below.

For example, with "rs45445694", click "Delete". The result is shown on the webpage below.

The Drug Suggestion aims to provide guidance for the effective management of specific diseases or conditions. This feature considers genetic variations and potential side effects, guiding users to identify appropriate medications based on their personal genetic profile, disease characteristics, and known efficacy and safety of drugs. The goal of medication recommendations is to provide personalized treatment plans by considering genetic factors that may influence drug response and potential adverse reactions. By utilizing genetic variation information and specific disease information, this section offers tailored medication suggestions for each user. The primary objective is to optimize treatment efficacy while minimizing the risks associated with medication use, ultimately improving patient outcomes.
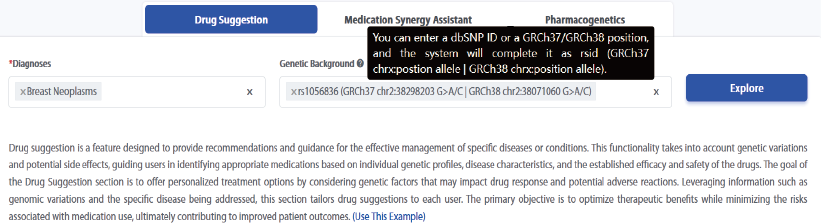
Users can search for diagnosed diseases or personal genetic backgrounds. Upon clicking 'Explore', a graph and summary will be provided, offering drug suggestions based on the entered information.
Genetic backgrounds provide representations in rsid (GRCh37 | GRCh38) format, such as rs1056836 (GRCh37 chr2:38298203 G>A/C | GRCh38 chr2:38071060 G>A/C). On our webpage, users can perform autocomplete searches using any part of these representations.
Additionally, tooltips have been added to the 'Genetic Background' section to explain that users can enter a dbSNP ID or a GRCh37/GRCh38 position, and the system will complete it as rsid (GRCh37 chrx:position allele | GRCh38 chrx:position allele).
The display of the webpage after clicking "Explore" includes:
Refer to the webpage below for details.
Note: Due to the input token limit of the underlying LLM, subgraph descriptions are limited to the top 10 knowledge items in the search results. To clarify, we have added the following tip on our webpage: " The following AI-generated knowledge descriptions are based on the top 10 results due to the token limit."

The Medication Synergy Assistant is designed to help individuals manage and optimize the combined use of multiple medications as well as the potential side effects of single drugs. It provides reminders and guidance to help users effectively and safely combine different medications, considering known effects and potential adverse reactions. The application takes into account the specific medications being used, including their expected therapeutic effects and possible side effects. It offers practice recommendations on the selection and use of medications to ensure maximum efficacy while minimizing the risk of harmful drug interactions or adverse reactions. The primary goal of the Medication Synergy Assistant is to enhance therapeutic outcomes and promote patient safety when using multiple medications, whether in combination or individually.
Users can search for specific drugs, drug combinations, or phenotypes of interest. Upon clicking 'Explore', a graph and recommendations for medication combinations will be displayed.

The display of the webpage after clicking "Explore" includes:
Refer to the webpage below for details.
Note: Due to the input token limit of the underlying LLM, subgraph descriptions are limited to the top 10 knowledge items in the search results. To clarify, we have added the following tip on our webpage: " The following AI-generated knowledge descriptions are based on the top 10 results due to the token limit."

Pharmacogenomics is the field that studies the impact of Genetic Background or specific Genes on individual drug responses. It involves analyzing how genetic factors influence drug metabolism, efficacy, and potential side effects. Pharmacogenomics explores the relationship between an individual's genetic makeup and the pharmacological mechanisms of drugs, aiming to provide genomic information related to specific medications. This includes identifying genetic variations or specific genes that may affect drug metabolism pathways, drug targets, or drug transport proteins. By understanding an individual's pharmacogenomic characteristics, researchers can gain insights into personalized drug responses and promote the development of targeted therapies.
Users can search for drugs, genes, or variations to receive pharmacogenomics-related recommendations.
Variations provide representations in rsid (GRCh37 | GRCh38) format, such as rs1056836 (GRCh37 chr2:38298203C>G | GRCh38 chr2:38071060G>C). On our webpage, users can perform autocomplete searches using any part of these representations.
Additionally, tooltips have been added to the 'Variation' section to explain that users can enter a dbSNP ID or a GRCh37/GRCh38 position, and the system will complete it as rsid (GRCh37 chrx:position allele | GRCh38 chrx:position allele).

The display of the webpage after clicking "Explore" includes:
Refer to the webpage below for details.
Note: Due to the input token limit of the underlying LLM, subgraph descriptions are limited to the top 10 knowledge items in the search results. To clarify, we have added the following tip on our webpage: " The following AI-generated knowledge descriptions are based on the top 10 results due to the token limit."
Users can click the "download" button to get the entity or pattern knowledge information.
