scMMO-atlas: single-cell multimodal omics atlas
A platform for displaying multiple omics of single-cell multimodal omics data
Welcome to scMMO-atlas
scMMO-atlas, accessible via
https://www.biosino.org/scMMO-atlas/
, serves as a comprehensive repository for single-cell multimodal omics data, covering gene expression, chromatin accessibility, proteins and a variety of other omics data.
This extensive resource comprises information from 3,168,824 human and mouse cells, spanning 27 cell lines/organs/tissues across 70 datasets.


Data summary
Distribution of species
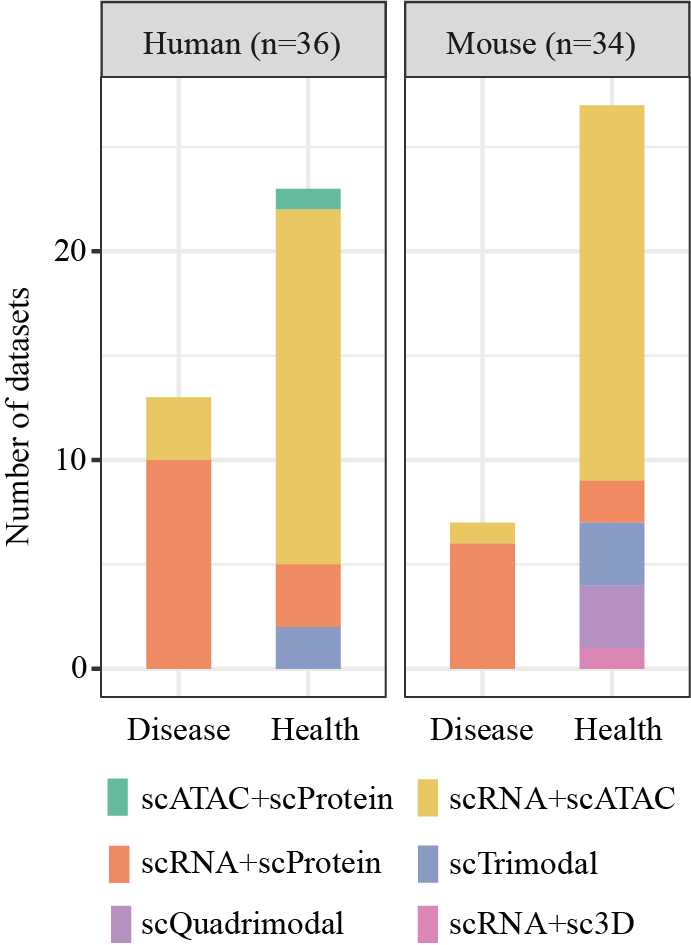
Distribution of doners condition and omics approach
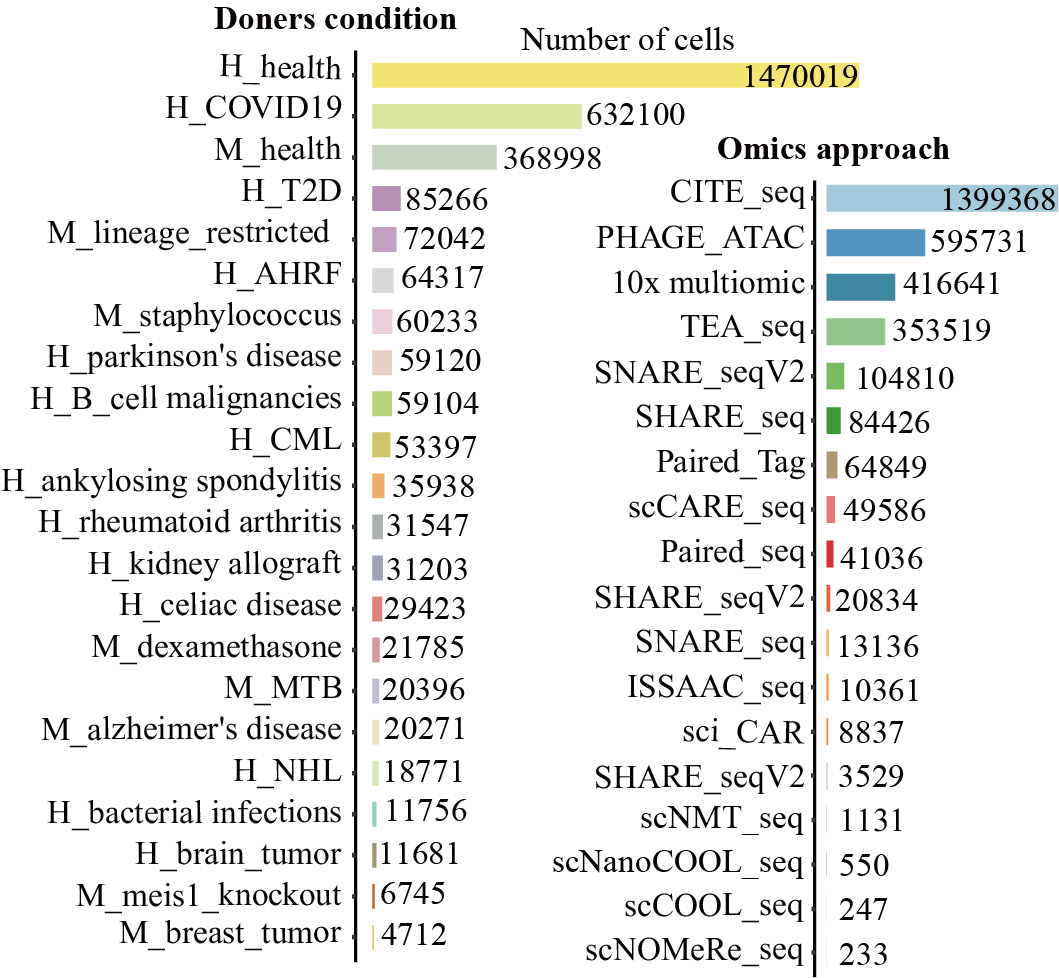
Distribution of tissues/organs
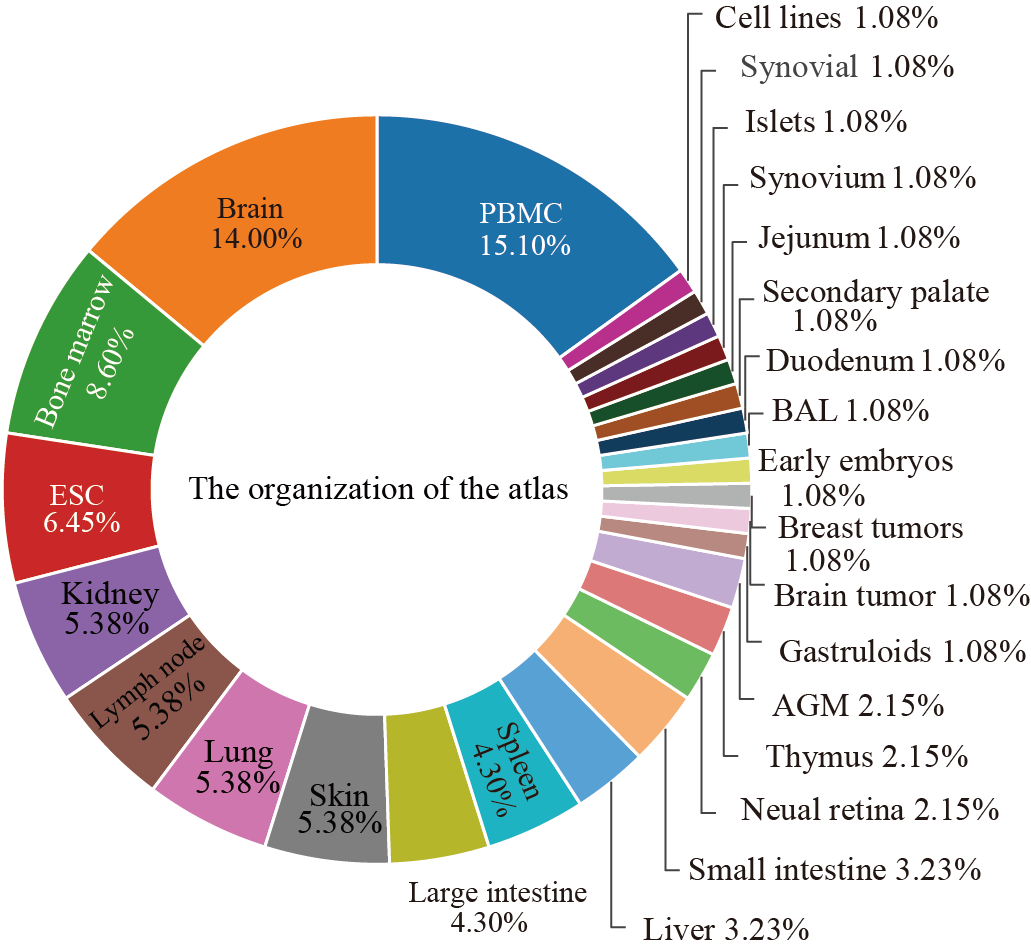
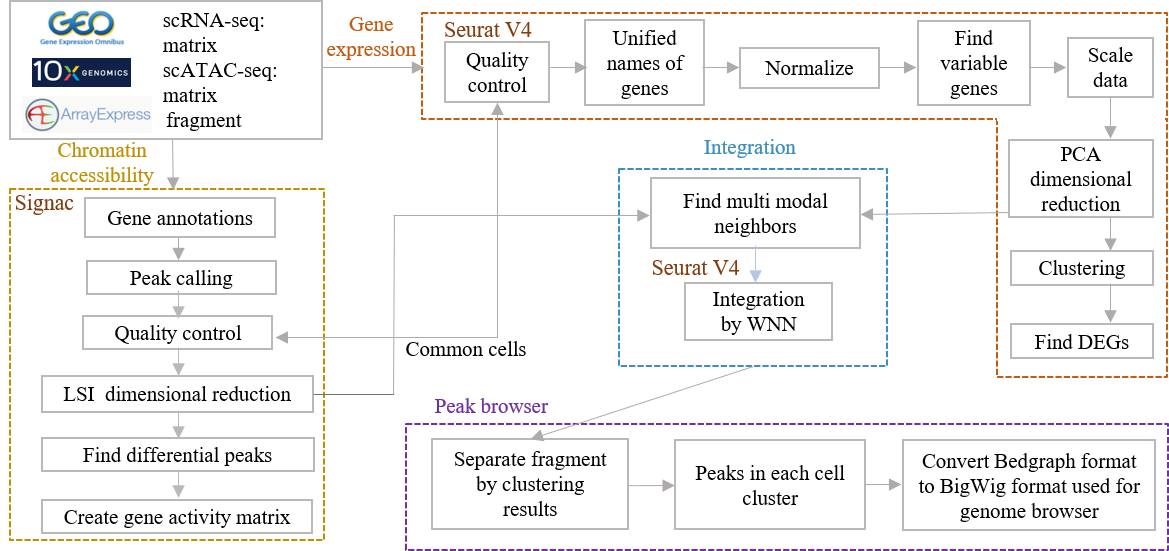
Database framework
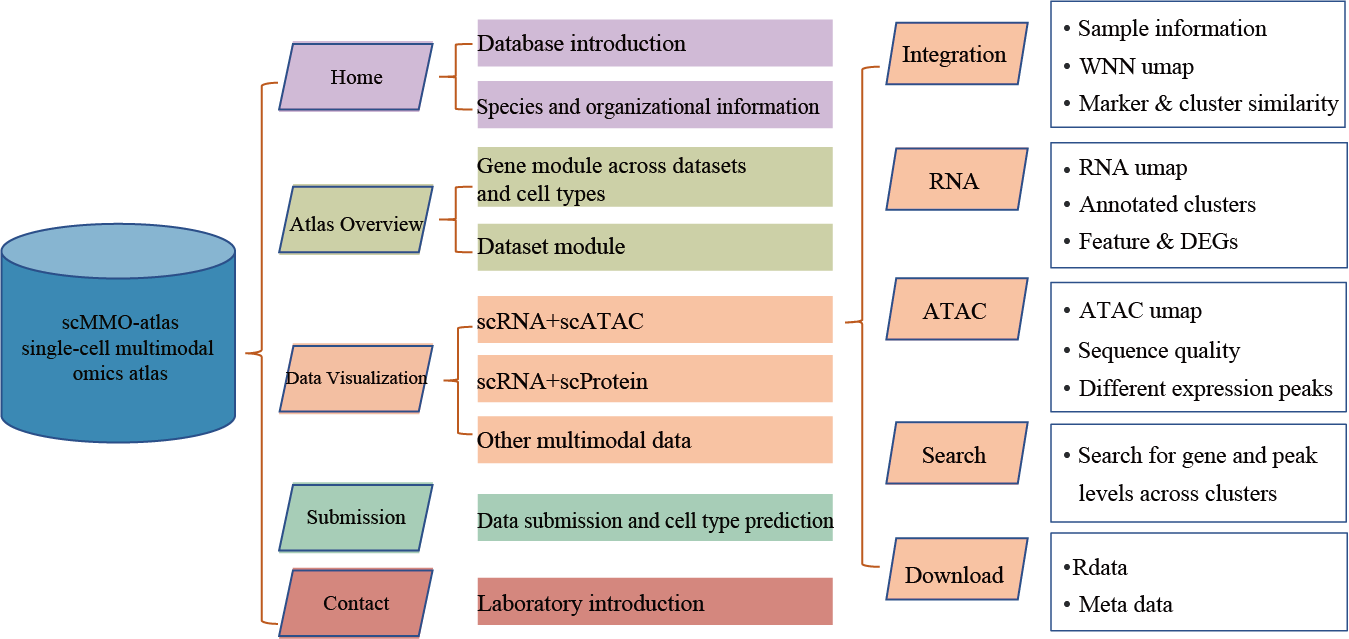
Data processing workflow (scRNA & scATAC data)
Protocol for data sharing in scMMO-atlas
If you would like to share your experimental data, please contact us via email: 12131301(a)mail.sustech.edu.cn. Please submit data in the following format:
1. The data needs to be stored in Rdata/Rds format.
2. For scRNA+scATAC data, please be sure to provide ATAC fragment and peak files.
Reference
Latest database update
Gene module
Loading...
Dataset module
Loading...
The
scRNA & scATAC
page offers comprehensive outcomes for annotated scRNA+scATAC data. Users begin by selecting their desired dataset through
Dropdown
, displaying sequencing approach, species and health, and cell number on the right. The
Integration
section showcases dataset specifics, an integrated UMAP plot with subgroup labels, highly expressed markers, and a corrlation heatmap.
RNA
and
ATAC
highlight data quality and subset-specific expressed genes/peaks. The
Search
section supports personalized exploration of gene expression in each cluster. The
Peak browser
allows gene-based or location-based enrichment per cell type. Annotated Rdata and metadata are accessible on the
Download
page.
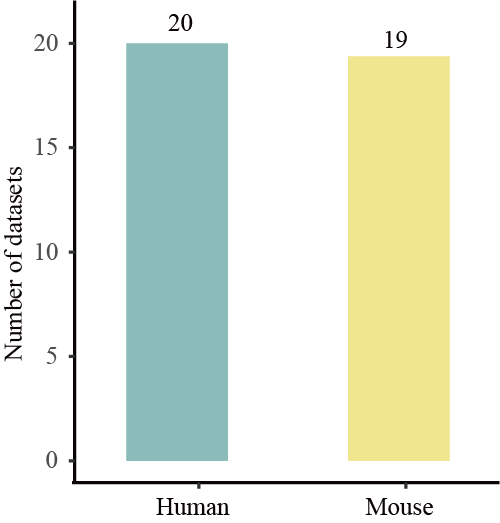
Data summary
Species of scRNA & scATAC data
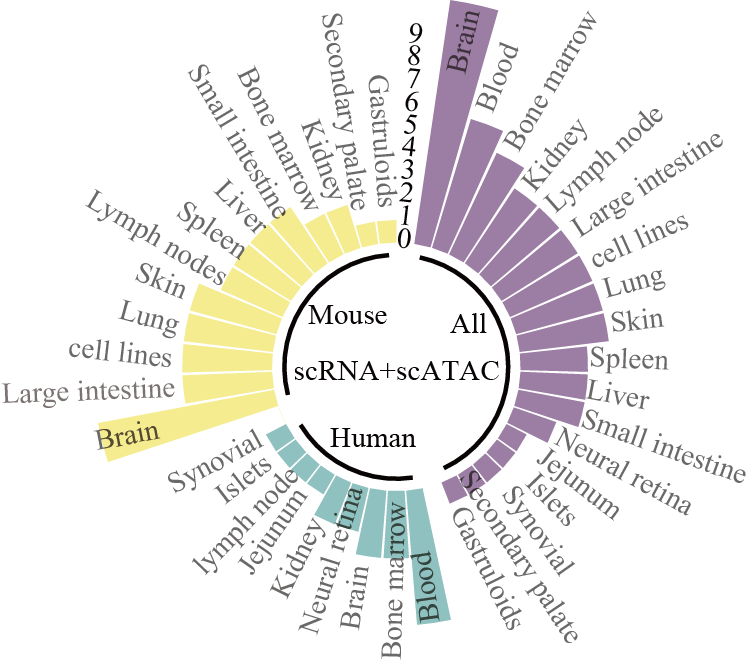
Tissues of scRNA & scATAC data
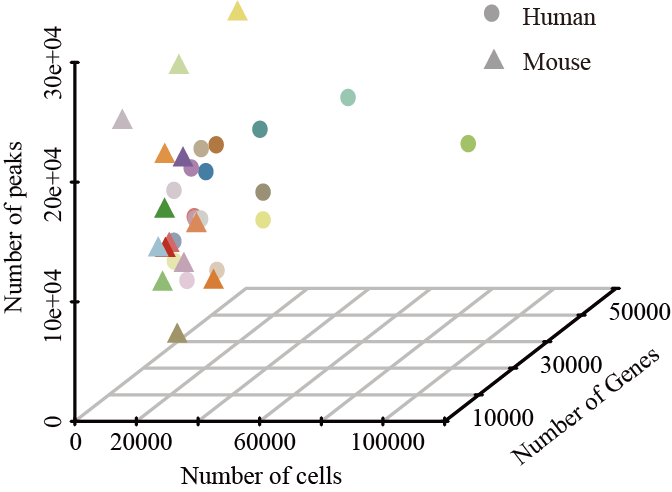
Summary of cells, genes and peaks
Loading...
Information
Loading...
Loading...
Peak browser
Download Metadata
Download RDS
scRNA and scProtein
Loading...
Loading...
Loading...
Loading...
Other single cell multimodal data types
Data set list
Loading...
Submission data and Prediction based on single-cell multimodal atlas
***Once submitted, your data will be saved on our server. We guarantee that the data will be used solely for research related to scMMO-atlas and will not be disclosed or made public.
Contact Us
We are dedicated to maintaining and enhancing this website, ensuring ongoing data updates. If you have any questions or suggestions, please don't hesitate to reach out to us. Your input is valuable to us.
Contact: Wenwen Cheng
Email: 12131301(a)mail.sustech.edu.cn
Adress: 1088 Xueyuan Blvd., Nanshan District, Shenzhen, China
Our lab: http://www.sinh.cas.cn/rcdw/qtyjzz/202312/t20231211_6908461.html
Copyright @
Lab of Dr. Wenfei Jin
of CAS Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences.