eLMSG Help
The eLMSG database is intended to integrate microbial systematics, genomics and phenomics (polyphasic taxonomy related phenotypes).
It will enclose information on taxonomy, ecology, morphology, physiology, and molecular
biology, etc. For approaching this aim, eLMSG was developed according to the following
scheme. In summary, there are three levels of data structures, corresponding to taxonomic
higher ranks (from phylum to genus), species and strains respectively.

For higher ranks level, the data content is divided into four categories mainly: General information, Description and emendation,
Subdivisions(s) and References. In addition, a Note part is displayed only when necessary.
The main data related to taxonomy, like the Latin name, taxon ID (NCBI taxonomy), etymology, rank, lineage, dates of effective and/or valid publications,
type (type strain list will be present for species rank), is organized in General
information part. The Subdivisions part include the records (entrances) of sub-taxa.
And the References part is a publication list that constituted the history of taxa
on taxonomy.
Note: Because in the current taxonomic system, some Latin names of taxa are identical to their sub-taxa, e.g. phylum Actinobacteria
and class Actinobacteria, we use a label <P> for phylum Actinobacteria for discriminating them.

For species level, besides General information and References, the data content also involve Ecology, Morphology, Physiology,
Biochemistry, Enzymology, Taxonomy marker, Genomics, and Strain list. All of
these parts try to present all the information about a species in a comprehensive way.

For strains level, besides some data in General information part, there is at least one genomic sequence data in Molecular
biology part. That means genomic data is obligatory to describe a strain in eLMSG.

For genomic data, all collected genomic sequences were re-annotated using same pipeline. Based on the results generated by
Glimmer3,
GeneMarkS, and
Prodigal, the pipeline prefers ORFs predicted by multiple methods firstly; and then
prefers ORFs with longest length; finally prefers ORFs (predicted by single method) with
domain information (Pfam). Functional annotation was based on the databases:
KEGG,
COGs,
Pfam,
Swiss-Prot,
TIGRFAMs,
GO, and
MetaCyc.

-
1. Simple search
The simple search allows for queries by entering a Latin name and/or a strain number. While entering the first letters, matching taxonomic names are suggested in a drop-down menu. -
2. Advanced search
The advanced search offers large-scale queries combining several data fields for comparative analyses of a multitude of species. Example: If you want to query the species that are positive for catalase in an appointed group like phylum Actinobacteria, you can make a combination of searching Actinobacteria in Lineage (General information) and searching ‘+’ (by selection) in catalase (Enzymology). -
3. Browser view
The Browser view offers the opportunity to access all available taxa by their taxonomic classification. Thereby you are able to browse through taxonomy and narrow down the list until you received the desired species.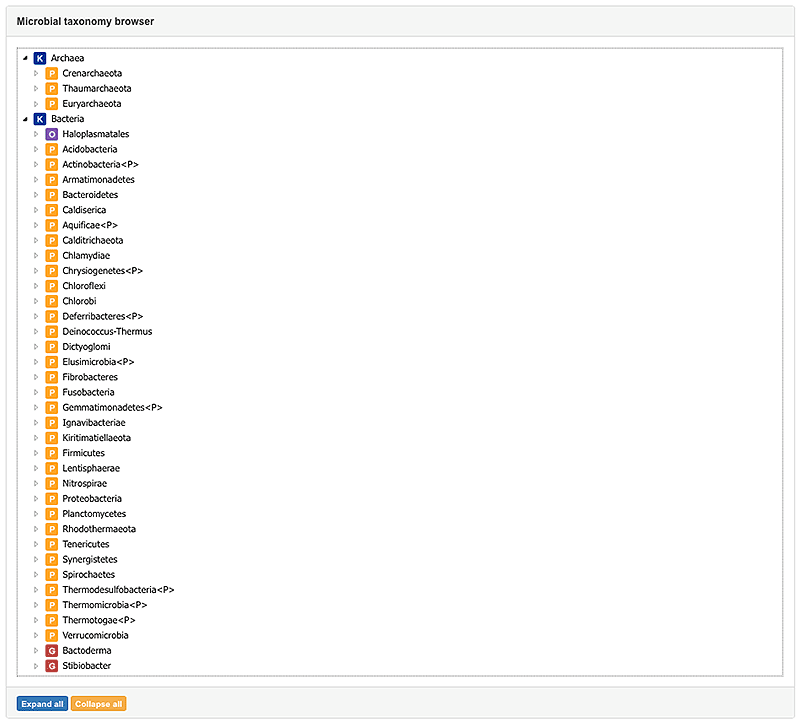
-
4. Data download
The sequence data including 16S rRNA and genome in eLMSG are free for downloading. When click download button, all genomic data (.fna, .frn, .faa, .gff, .rRNA, .tRNA, .ko, .cog, .pfam, .sprot, .tigr, .ec, .cyc, .go, and/or a .pan file of pangenome analysis result, if the species have more than 5 strains with genomic sequences) will be compressed as a tar.gz file for downloading. -
5. Statistics
The Statistics page presents a brief summary of data included in current version of eLMSG system. -
6. Analysis
The current eLMSG provides an analysis service for identifying the 16S rRNA sequence based on sequence similarity. The reference database of 16S rRNA sequence is quality-controlled and enclosed the sequences obtained by PCR amplification and/or genome next-generation sequencing.
Output Example:
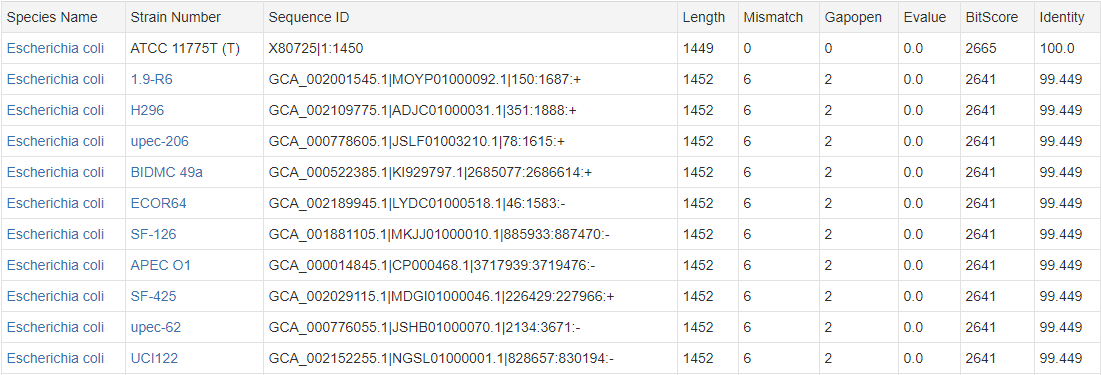
There are two kinds of Sequence ID, corresponding to two sequence acquisition methods: a, Accession|Start:End, example: X80725|1:1450, for sequences obtained by PCR amplification; b, AssemblyID|ScaffoldID|Start:End:Strand, example: GCA_002001545.1|MOYP01000092.1|150:1687:+, for sequences obtained by genome sequencing.
