Help
Cancer Type Abbreviation
| Abbreviation | Cancer type | Abbreviation | Cancer type | |
| 1 | ADCA | Adrenocortical Cancer | BTCA | Biliary Tract Cancer |
| 2 | BLCA | Bladder Cancer | BNCA | Brain Cancer |
| 3 | BRCA | Breast Cancer | CECA | Cervical Cancer |
| 4 | CHOR | Chordoma | COAD | Colon Cancer |
| 5 | CLCA | Colorectal Cancer | ENCA | Endometrial Cancer |
| 6 | ESCA | Esophageal Cancer | GBCA | Gallbladder Carcinoma |
| 7 | GSCA | Gastric Cancer | GAST | Gastrointestinal Stromal Tumor |
| 8 | HNSC | Head and Neck Cancer | HEGI | Hemangioma |
| 9 | LIHC | Hepatocellular Carcinoma | KDCA | Kidney Cancer |
| 10 | LNCA | Larynx Cancer | LEUK | Leukemia |
| 11 | LICA | Liver Cancer | LUCA | Lung Cancer |
| 12 | LYMP | Lymphoma | MELA | Melanoma |
| 13 | MESO | Mesothelioma | NSCA | Neuroendocrine Neoplasia |
| 14 | NDCA | Neuroendocrine Neoplasia | OPSC | Oropharyngeal Squamous Cell Carcinoma |
| 15 | OSCA | Oral Squamous Cell Carcinoma | OVCA | Ovarian Cancer |
| 16 | PNCA | Pancreatic Cancer | PRCA | Prostate Cancer |
| 17 | RETI | Retinoblastoma | SCRA | Sarcoma |
| 18 | SKCA | Skin Cancer | SINT | Small Intestinal Neuroendocrine Tumor |
| 19 | TECA | Testicular Cancer | THCA | Thyroid Cancer |
| 20 | TOCA | Tonsil Cancer | UTCA | Uterus Cancer |
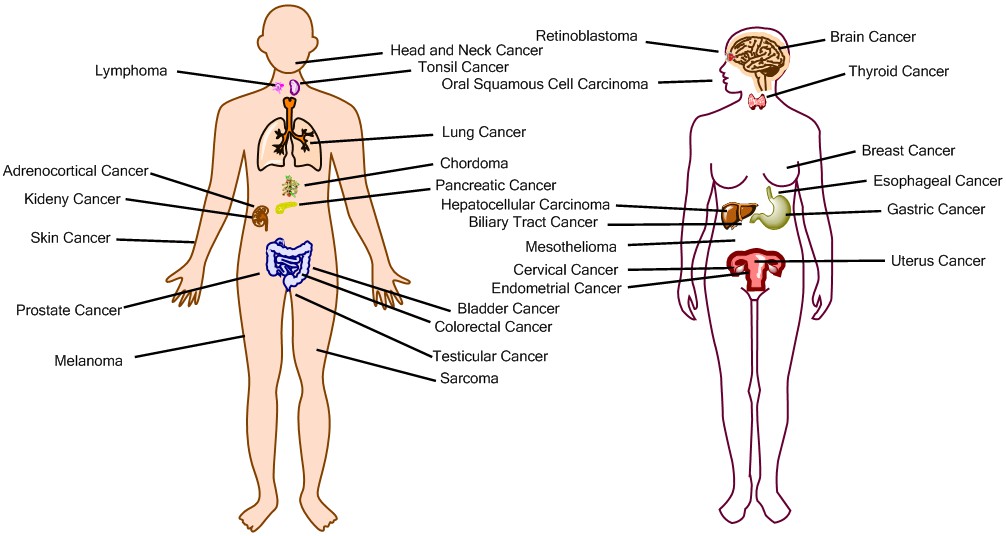
Schematic Diagram for Cancer Location
Schematic Diagram for the location of major cancer types included in database
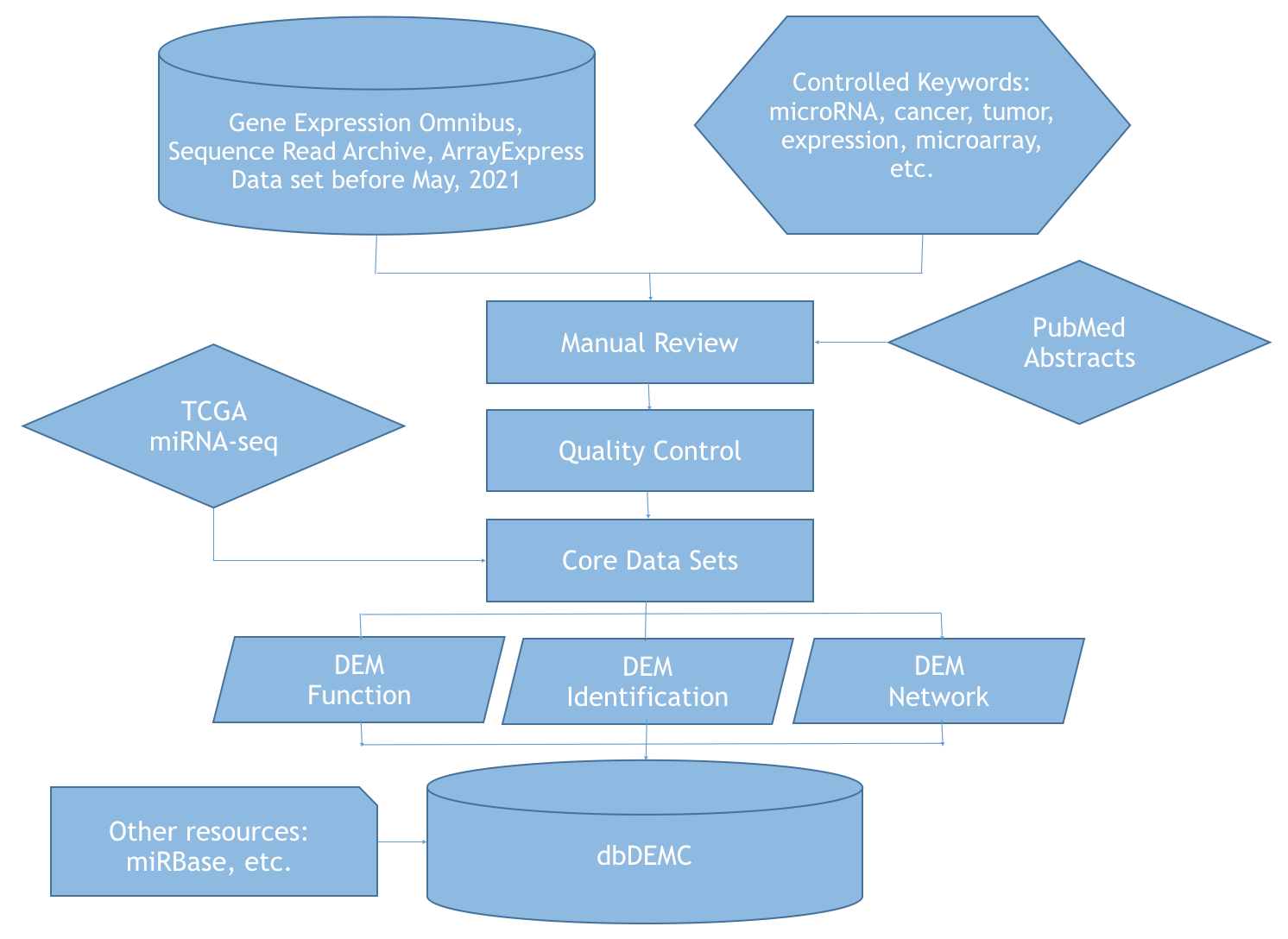
Data Analysis Flowchart
All differentially expressed miRNAs (DEMs) in this database are curated from public resources. The following work flow demonstrates the data collection process:
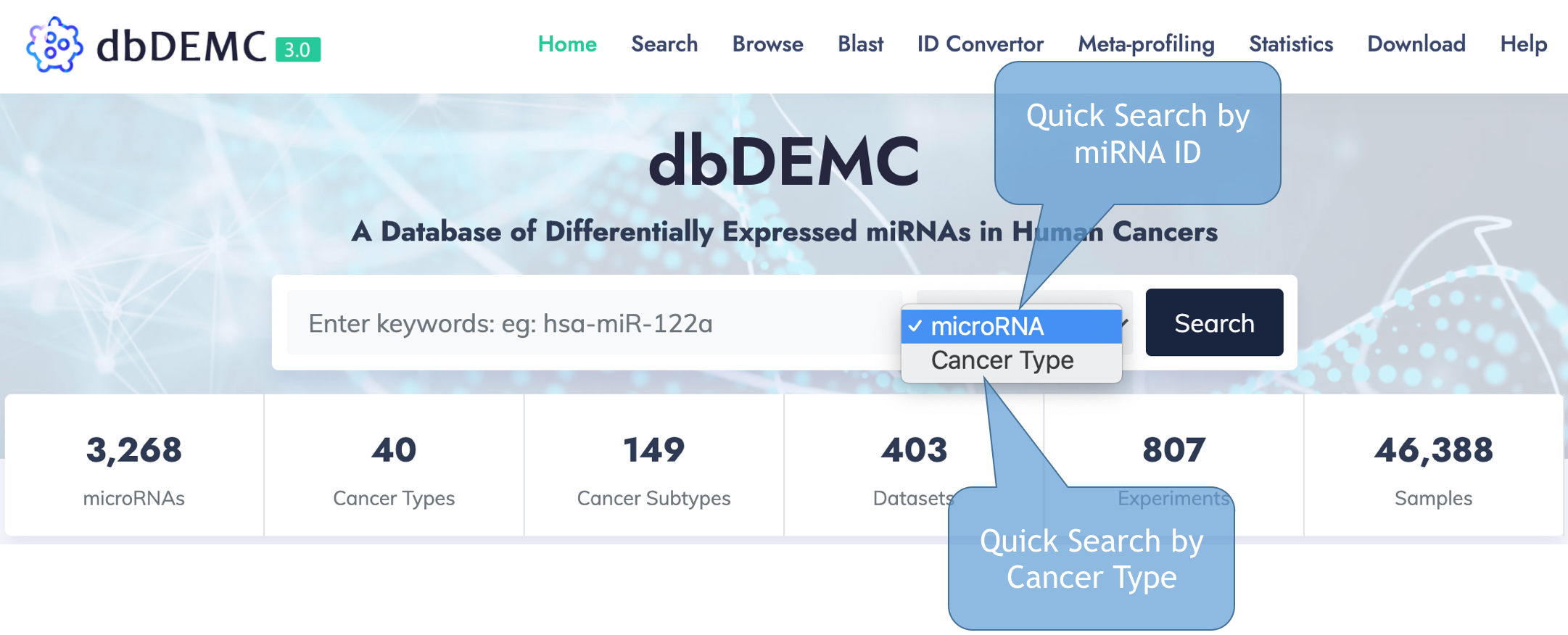
Database Search
dbDEMC allows for quick search by miRNA ID or specific cancer name from main page.
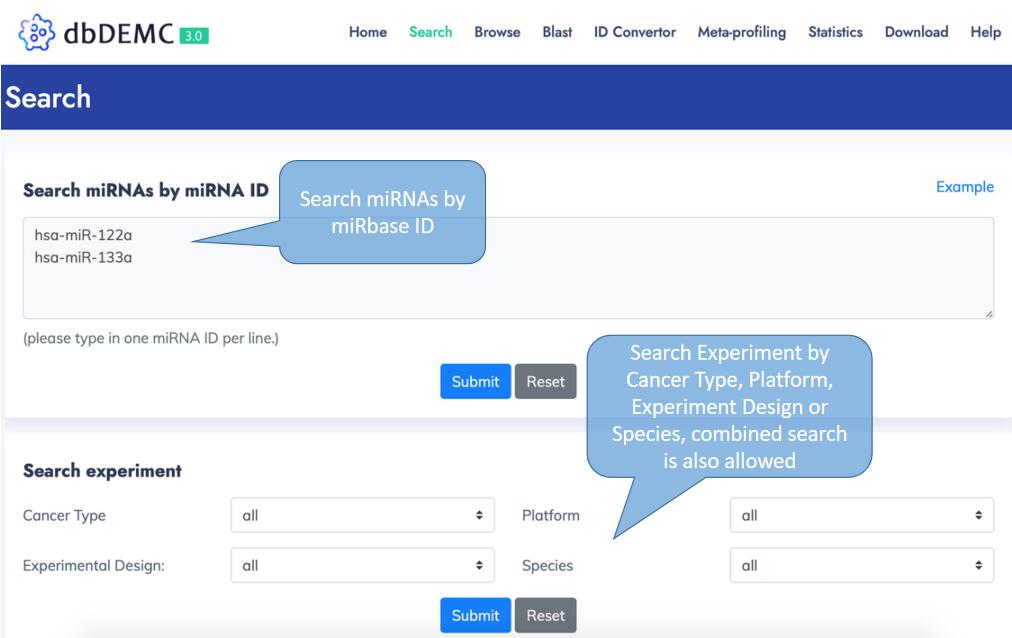
You can also make a detail search by miRNA IDs or search experiments by particular filter criteria from Search page.
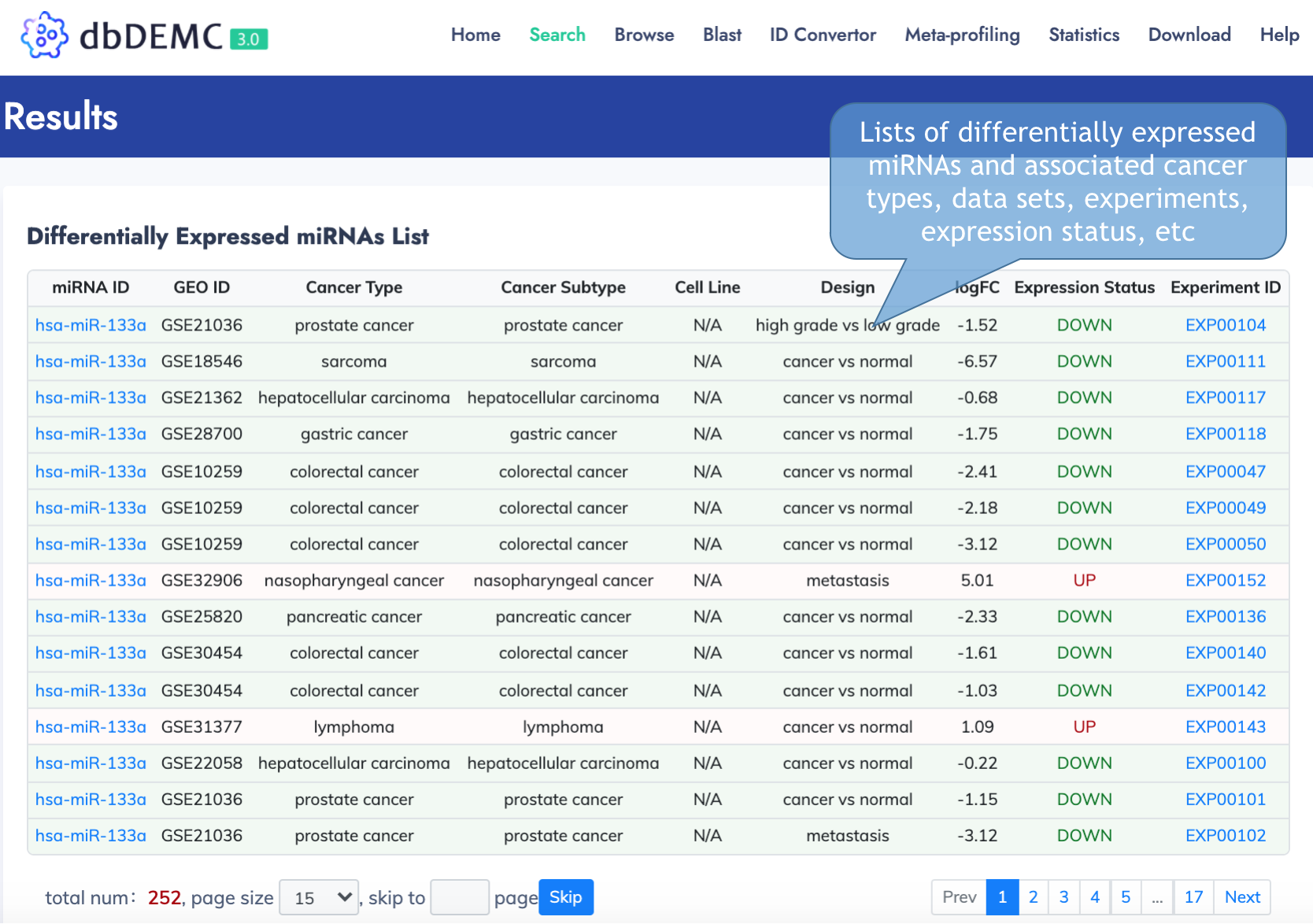
Search result page
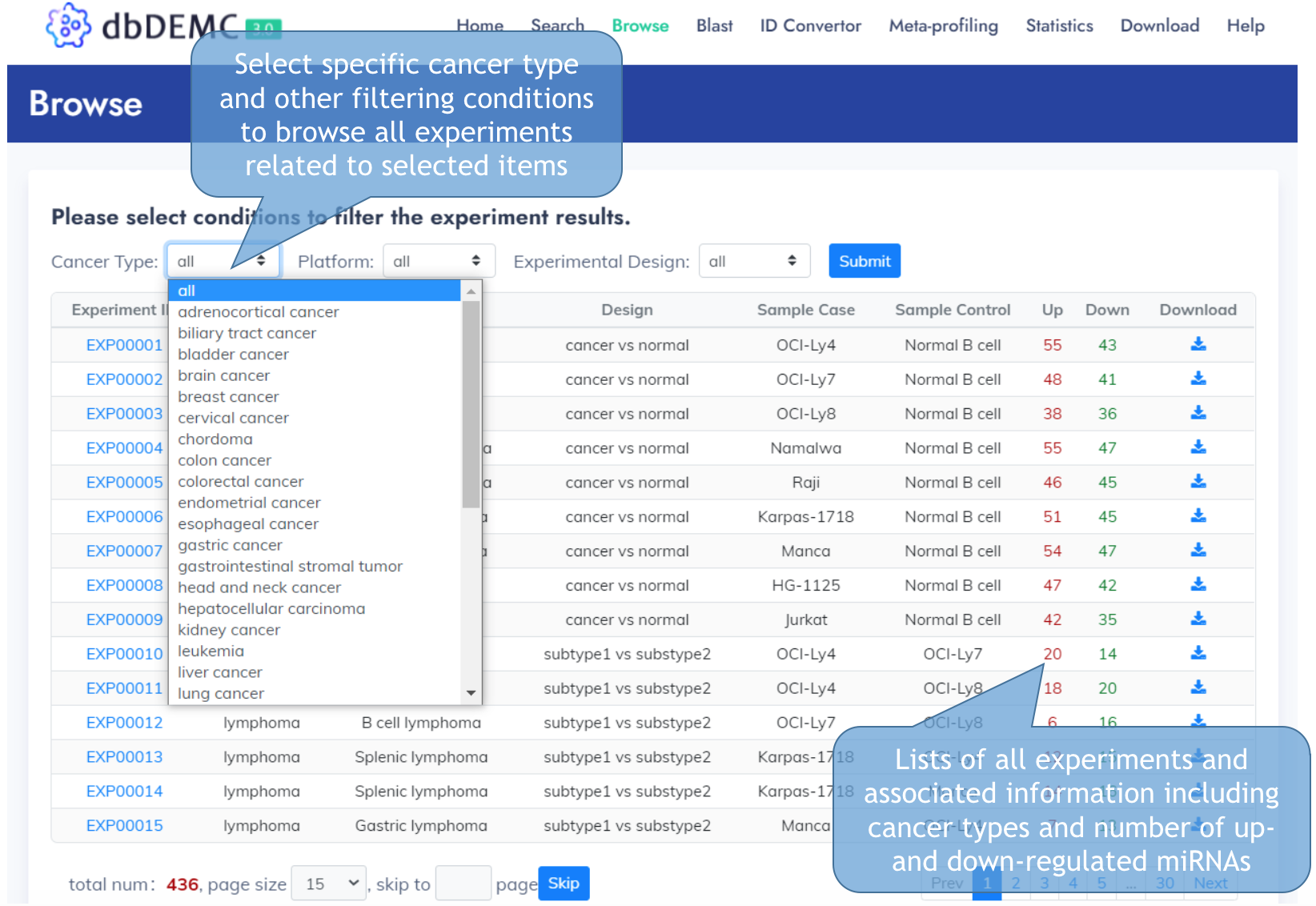
Browse Data
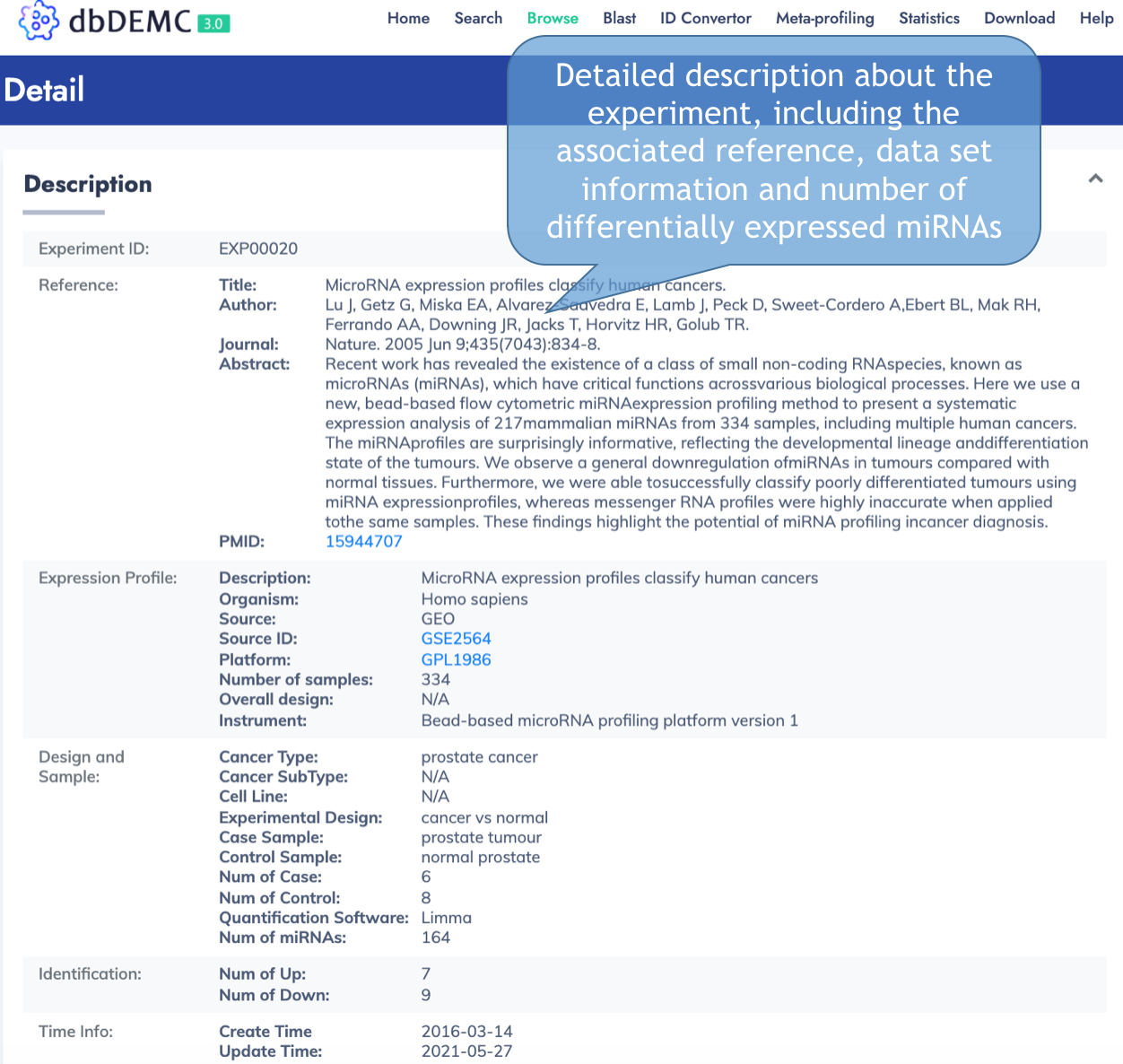
Experiment Detail
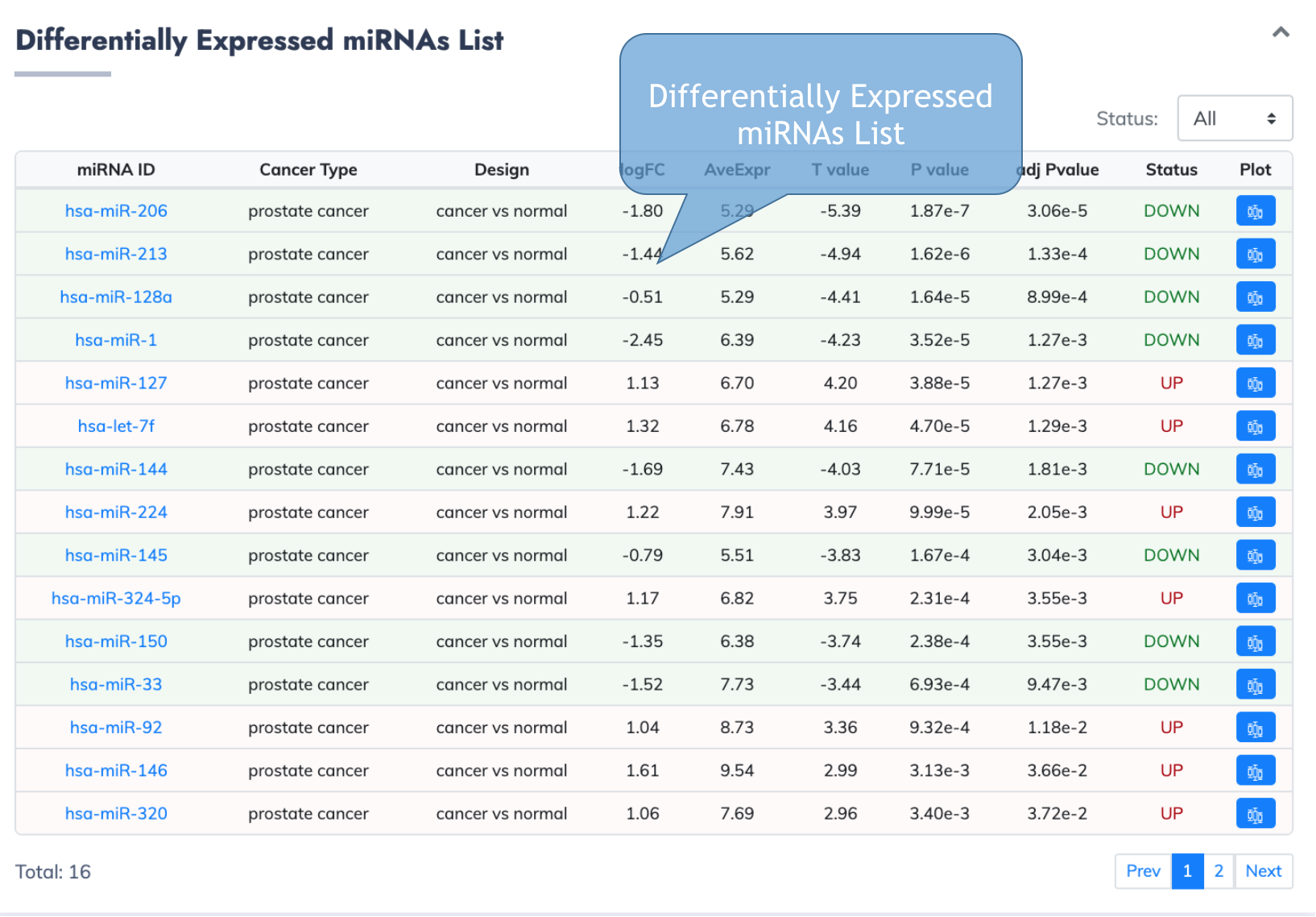
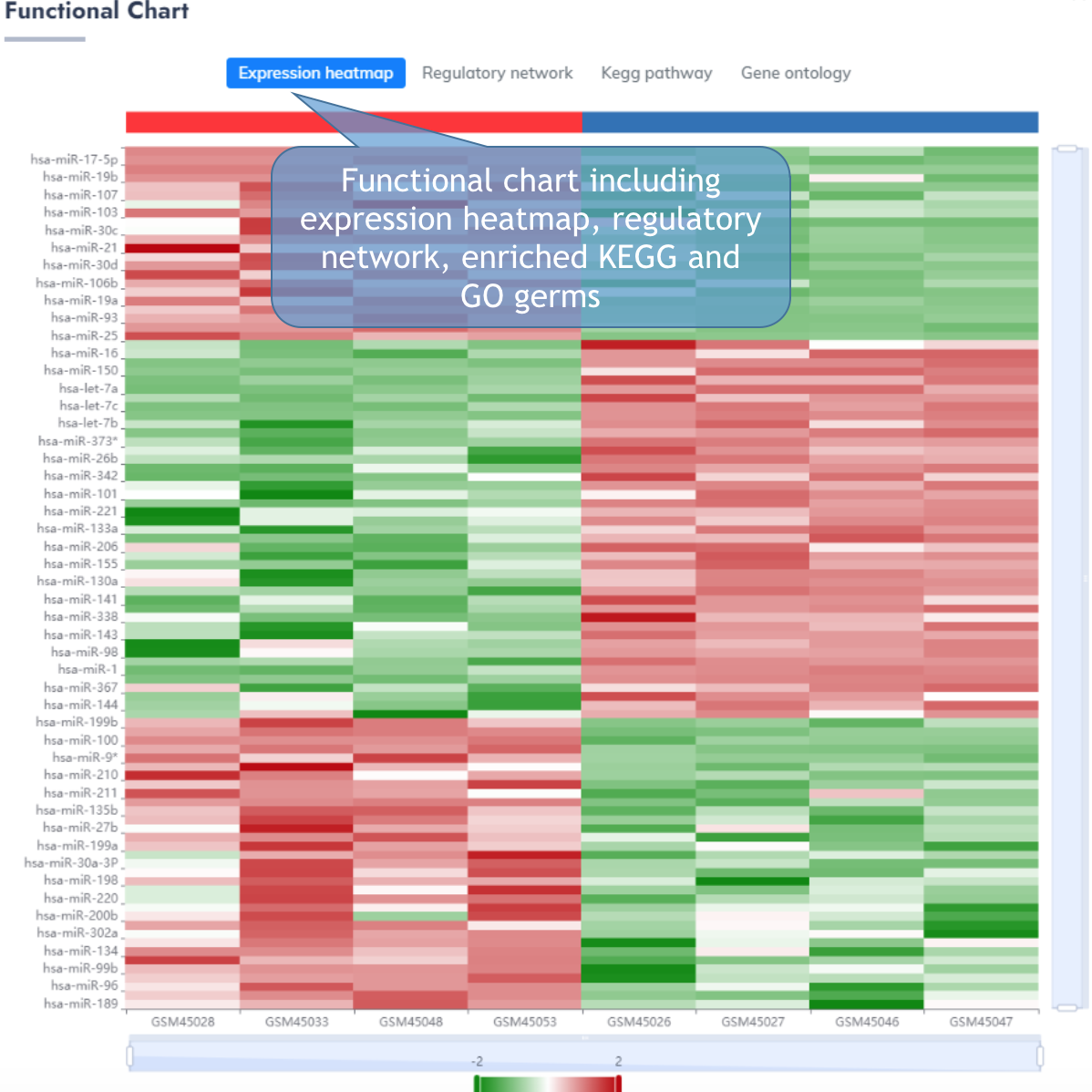
MiRNA Information Page
This page delineates the detailed information for each differentially expressed miRNAs.
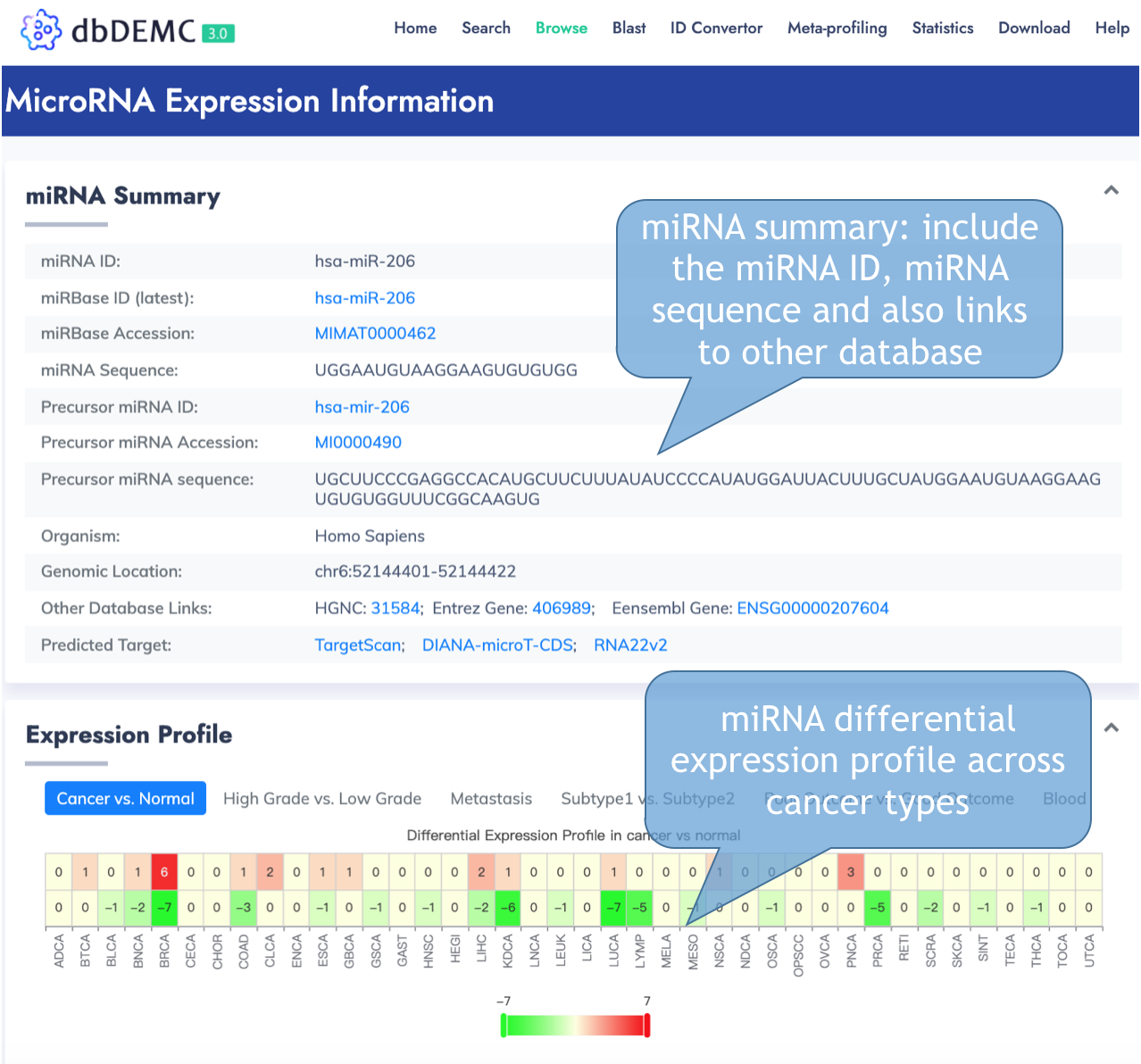
Detailed expression information from limma are listed.
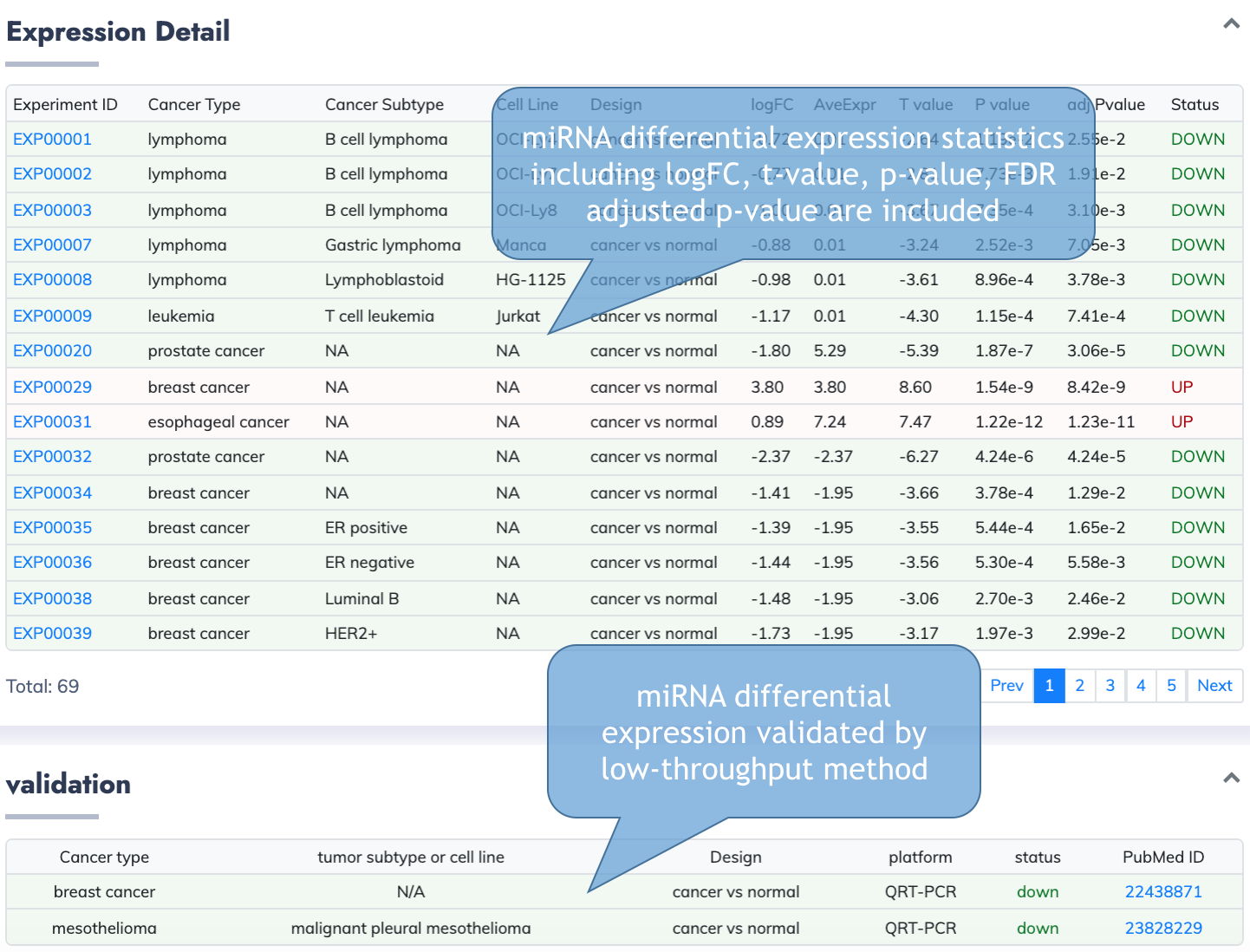
ID convertor
Convert miRBase old version ID to miRBase latest version ID
Meta-profiling
You can input miRNA IDs and view the Meta-profiling heatmap
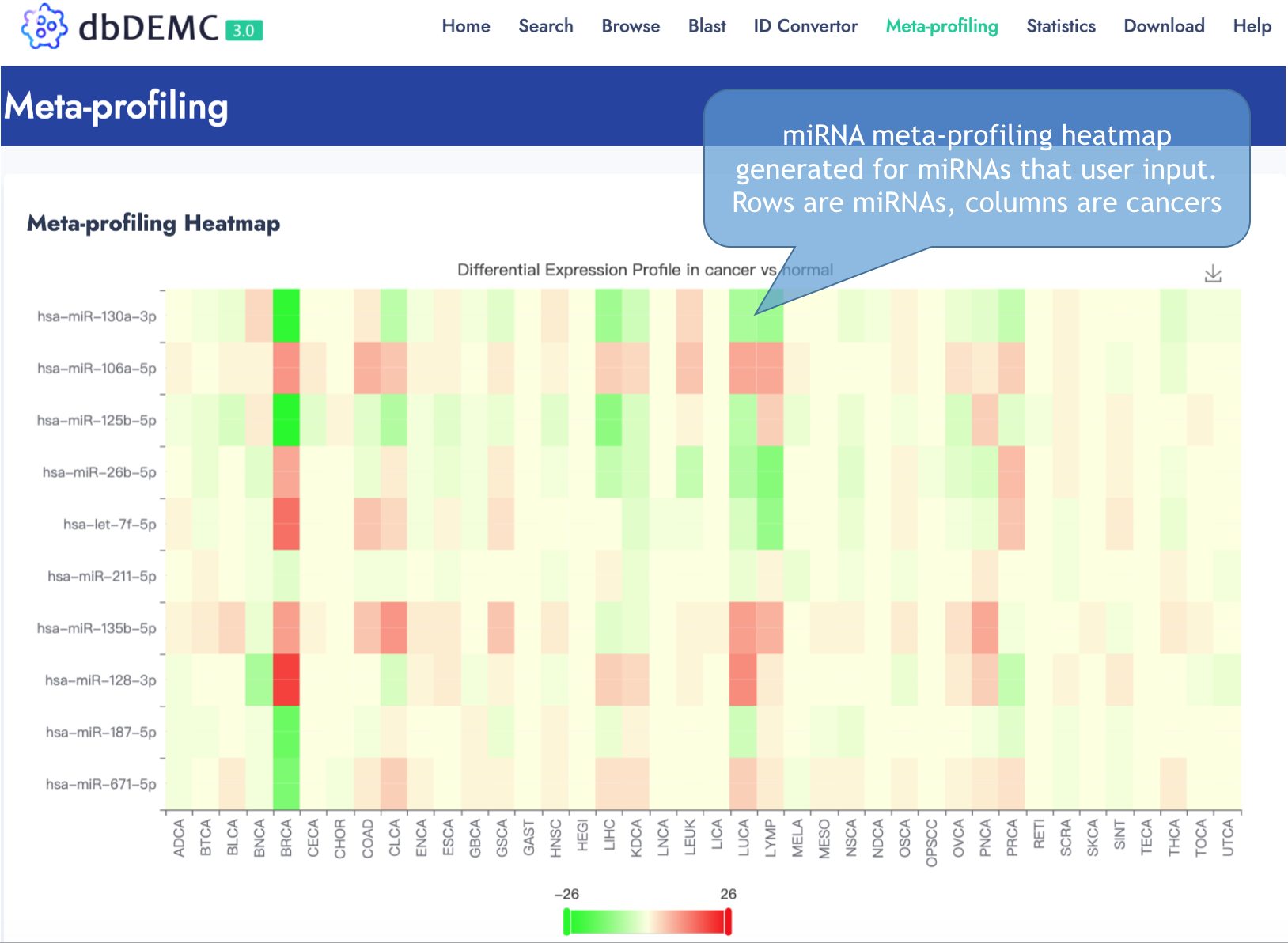
Example of Meta-profiling heatmap
Publications
[1]Feng Xu, Yifan Wang, Yunchao Ling, et al. "dbDEMC 3.0: Functional exploration of differentially expressed miRNAs in cancers of human and model organisms". Genomics, Proteomics & Bioinformatics. 2022; doi: https://doi.org/10.1016/j.gpb.2022.04.006 [2]Zhen Yang, Liangcai Wu, Anqiang Wang, et al. "dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers". Nucleic Acids Research. 2017; 45(D1):D812-D818. [3]Zhen Yang, Fei Ren, Changning Liu, et al. "dbDEMC: a database of differentially expressed miRNAs in human cancers". BMC Genomics. 2010; 11(Suppl 4): S5.About Us
This database is hosted by Computational Genetics Group, Institute of Biomedical Sciences, Fudan University
Contact Us:
- 138 Medical College Road, Fenglin Campus of Fudan University,Shanghai
- Dr. Zhen Yang