1. Get an overview of GTDB on home page.
5. BLAST search
7. GTdock
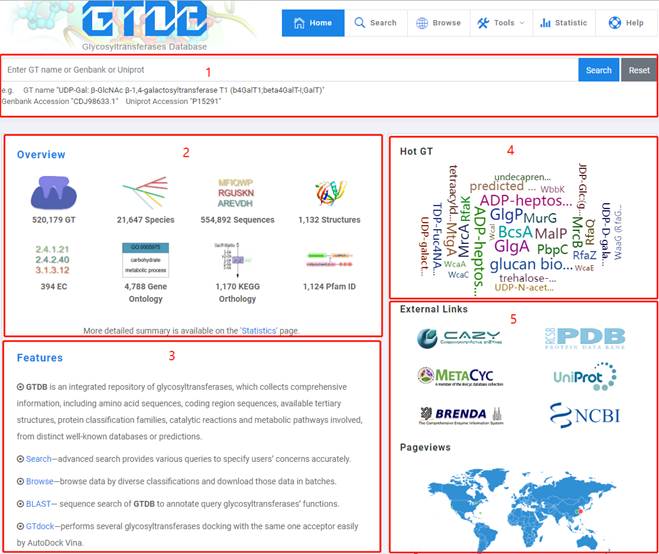
1. Get an overview of GTDB on home page.
1. You can input "GT name" or "Genbank Accession" or "Uniprot Accession" to have a quick view in the simple search bar. And it's noticeable that fuzzy match is only used in search of "GT name" while perfect match is used in search of "Genbank Accession" and "Uniprot Accession".
2. An overview of mainstream data in GTDB.
3. Features and key functions of GTDB.
4. You can click on any of hot GT in word cloud to browse corresponding details.
5. External links and pageviews are provided.
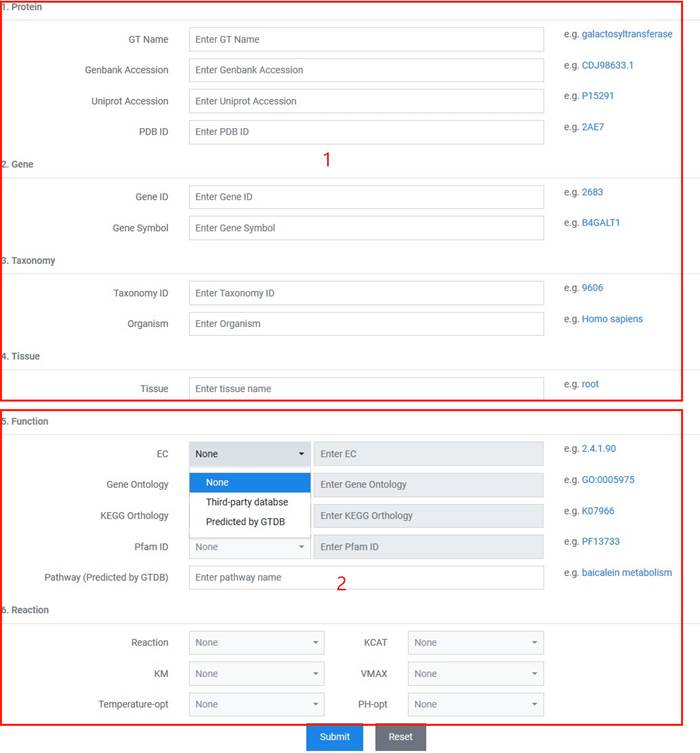
1. You can input various query items that meet the requirements of fuzzy match and boolean operator (only "and").
2. "Third party database" or "Predicted by GTDB" method options are provided for searching GT functions and reactions.
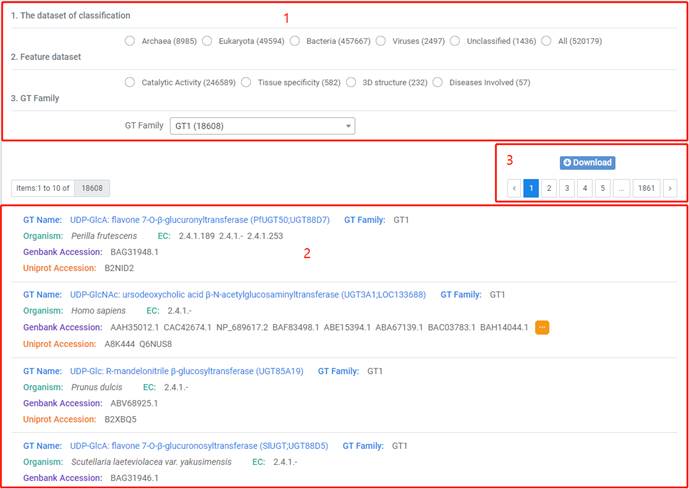
1. You can browse data by taxonomy, GT family and other features.
2. You can click on "GT name" to view details.
3. You can click on "Download" button to download the annotations of selected dataset.
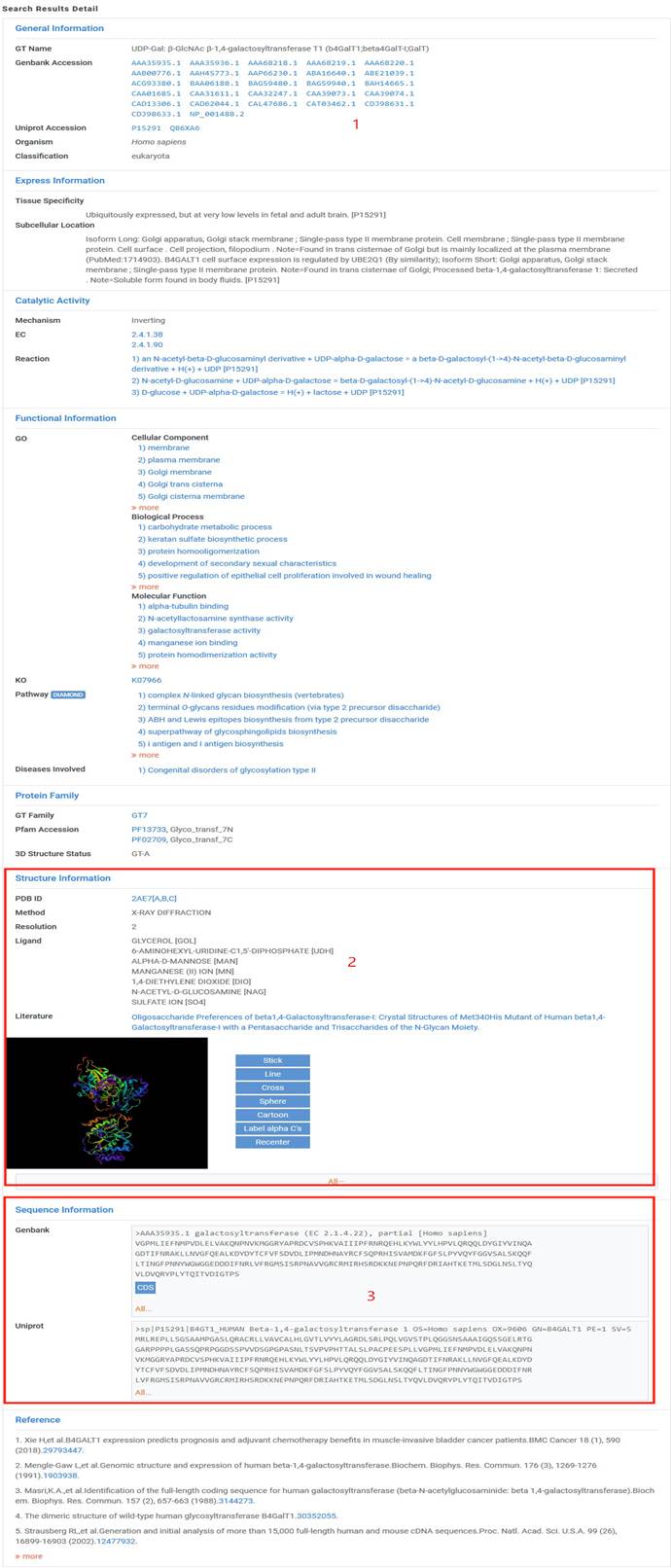
1. In the search detail page, you can get information including General Information, Express Information, Catalytic Activity, Functional Information, Structure Information, Sequence Information and Reference.
2. Click on button "All" in "Structure Information" , one new tab will show all structures of this entry.
3. Click on button "All" in "Sequence Information" , one new tab will show all protein sequences of this entry and "CDS" will display the corresponding coding sequence. Moreover, click on the "FASTA" button in new tab, you can download all protein and coding sequences of this entry.
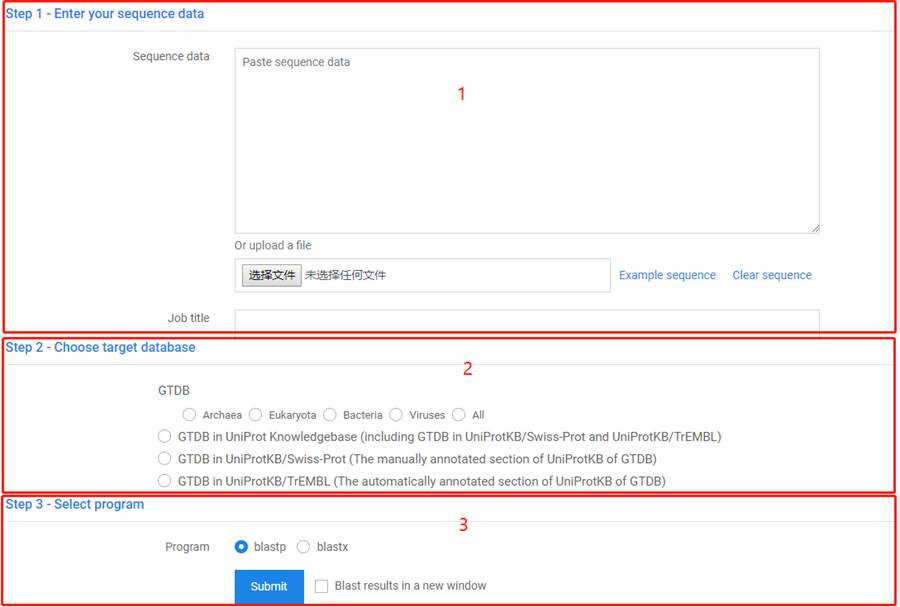
1. You can paste or upload less than 10 sequences.
2. You can choose target database by taxonomy or by different identification ways.
3. Select program according to input sequence.
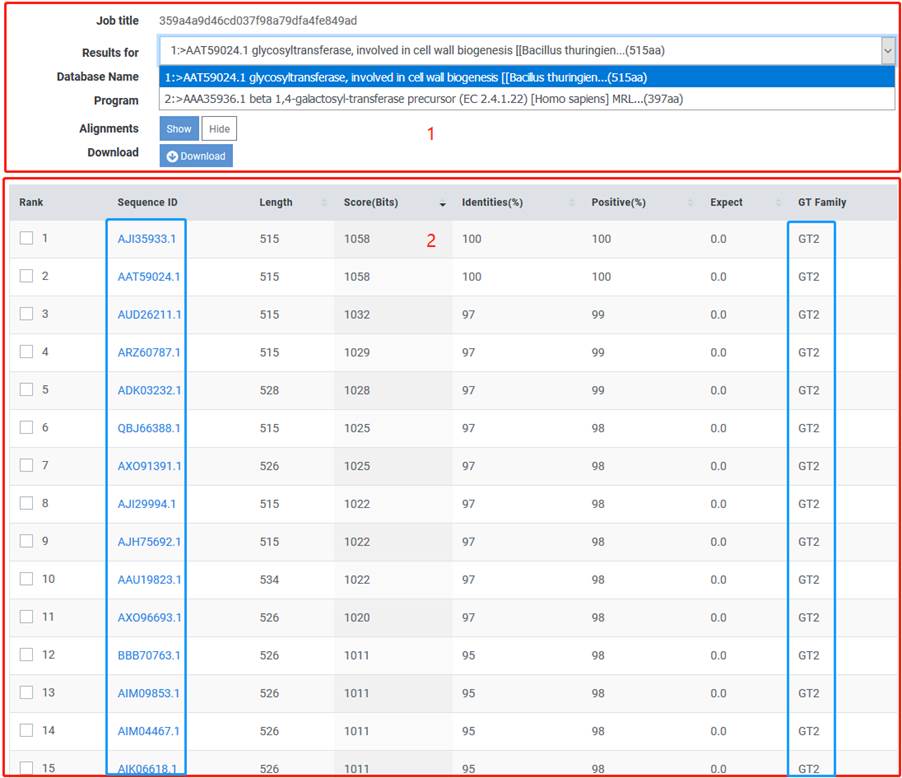
1. You can click the drop-down to select the submitted sequence and click on the "Download" button to download the corresponding sequence results.
2. You can view detail information of similar sequence to help for understanding the query sequence.
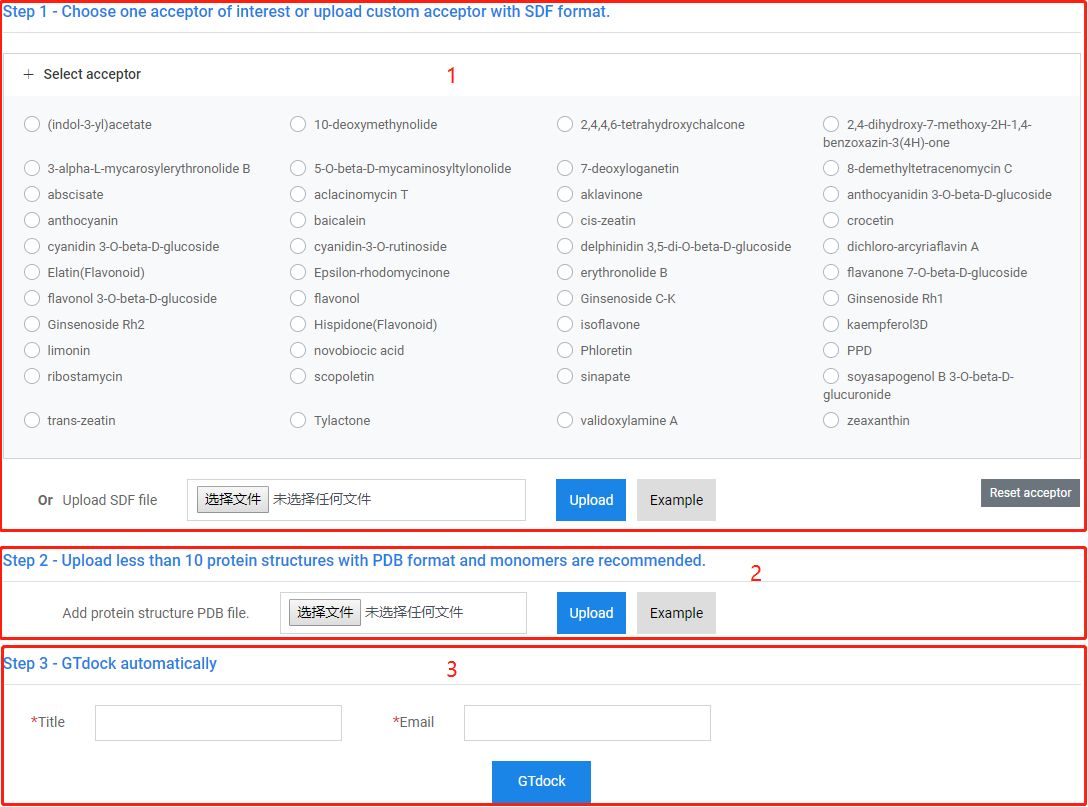
1. Choose one acceptor or upload a custom acceptor.
2. Upload less than 10 proteins and monomers are recommended.
3. Fill in the job title and your email, then you can start GTdock.
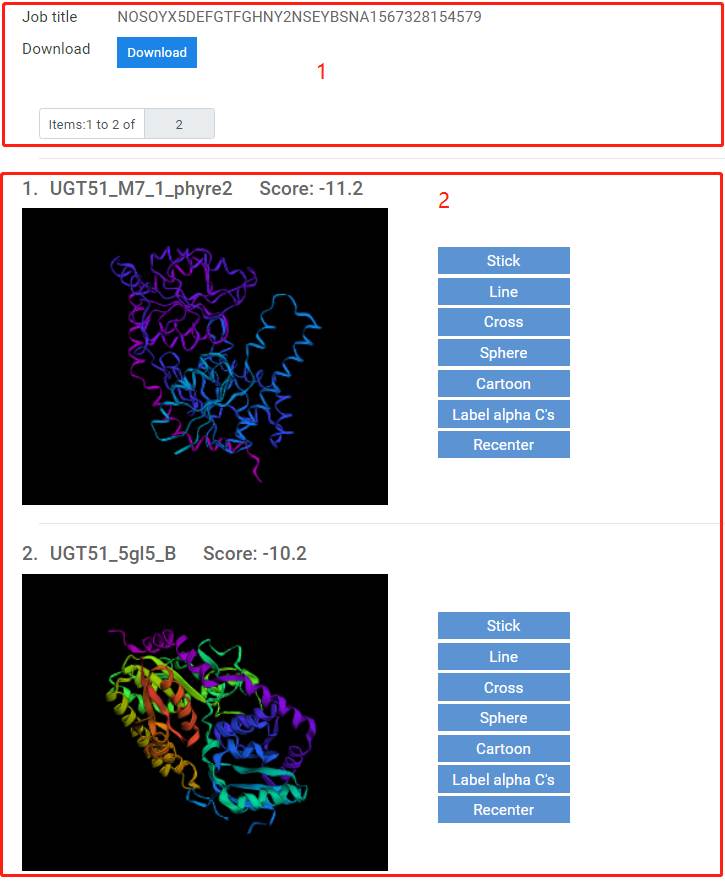
1. You can directly download the GTdock results, including BLAST results, scores of proteins and all docking results with PDB format.
2. You can view the GTdock results on our web page.