A demo data was imputed using the Han Deep Sequencing reference panel. In total, data of 10 individuals with 29,511,084 variants were generated. Results in Plink binary format and VCF format can be downloaded.
Summary:
Genotype imputation, or simply imputation in the context of our database, is to estimate the unobserved genotypes in the given dataset. Our imputation service is designed to achieve the best imputation result of Han population data with a reference panel based on our multiple datasets of Han Chinese.
Our imputation service is implemented by commonly used tools: SHAPEIT4, and IMPUTE5. There are 9 reference panels available in our imputation service. In PGG Han 2.0, the panel of Han deep sequencing data and six Han regional substructure population data are newly introduced. Currently, the imputation function is limited to the biallelic SNV data.
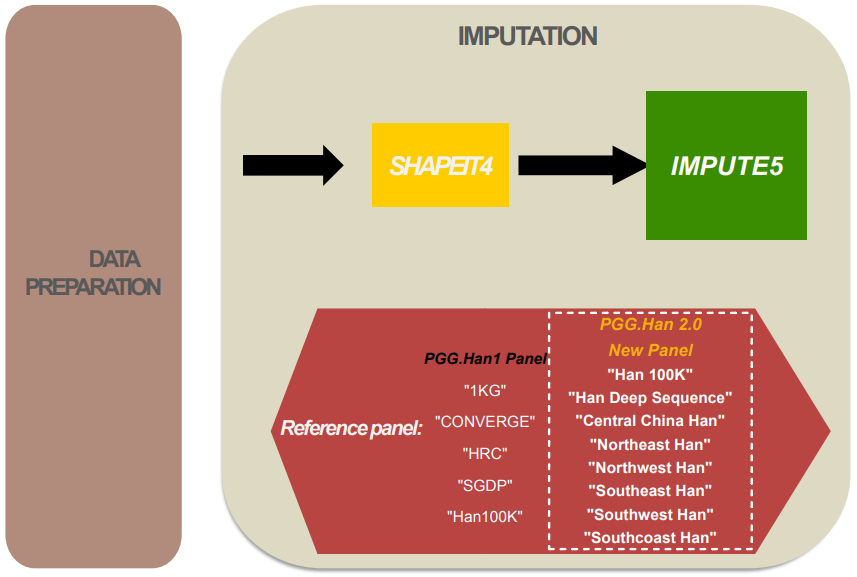
Software and references:
SHAPEIT4
Delaneau, O., Zagury, J.-F., Robinson, M.R., Marchini, J., and Dermitzakis, E. (2018). Integrative haplotype estimation with sub-linear complexity. BioRxiv 493403.
IMPUTE5
Rubinacci S, Delaneau O, Marchini J. Genotype imputation using the Positional Burrows Wheeler Transform. PLoS Genet. 2020 Nov 16;16(11):e1009049. doi: 10.1371/journal.pgen.1009049. PMID: 33196638; PMCID: PMC7704051.
Reference panels:
CONVERGE
Reference panel based on CONVERGE dataset which only keeps the sites passed the filter recommended by the author of the paper “11,670 whole-genome sequences representative of the Han Chinese population from the CONVERGE project”(10,640 Han females, 5,814,870 variants)
Han100K
A reference panel of highly representative core Han Chinese genomes covering all six genetic substructure regions of Han Chinese was selected from previous genotyping or partially imputed dataset Han100K (102,586 individuals, 5,042,439 variants).
Han deep sequencing referencece panel high-quality samples selected from multiple Han datasets (17,615 individuals, 160,992,704 variants).
Subgroup Central China HanA reference panel of highly representative core Han Chinese genomes covering Subgroup Central China Han selected from Han100K reference panel (1000 individuals, 5,042,439 variants).
Subgroup Northeast Han
A reference panel of highly representative core Han Chinese genomes covering Subgroup Northeast Han was selected from the Han100K reference panel (1000 individuals, 5,042,439 variants).
Subgroup Northwest Han
A reference panel of highly representative core Han Chinese genomes covering Subgroup Northwest Han was selected from the Han100K reference panel (1000 individuals, 5,042,439 variants).
Subgroup Southeast Han
A reference panel of highly representative core Han Chinese genomes covering Subgroup Southeast Han has been selected from the Han100K reference panel (1000 individuals, 5,042,439 variants).
Subgroup Southwest Han
A reference panel of highly representative core Han Chinese genomes covering Subgroup Southwest was selected from the Han100K reference panel (1000 individuals, 5,042,439 variants).
Subgroup South Coast Han
A reference panel of highly representative core Han Chinese genomes covering Subgroup South Coast Han was selected from the Han100K reference panel (1000 individuals, 5,042,439 variants).