xMarkerFinder is a cutting-edge computational framework designed to unravel the complexities of microbial biomarkers. At the heart of its mission lies the aim to find universally robust biomarkers in clinical and environmental settings.
Biomarker discovery in real-world scenarios presents two primary challenges. The first involves eliminating data heterogeneity across diverse cohorts to ensure robust findings. The second entails leveraging available retrospective data, often in abundance, to compensate for the frequently limited prospective data in a majority of situations. Here, xMarkerFinder effectively addresses these two challenges by employing meta-analysis techniques and introducing the specificity assessment with non-target datasets.
Take a look at the four stages of xMarkerFinder!
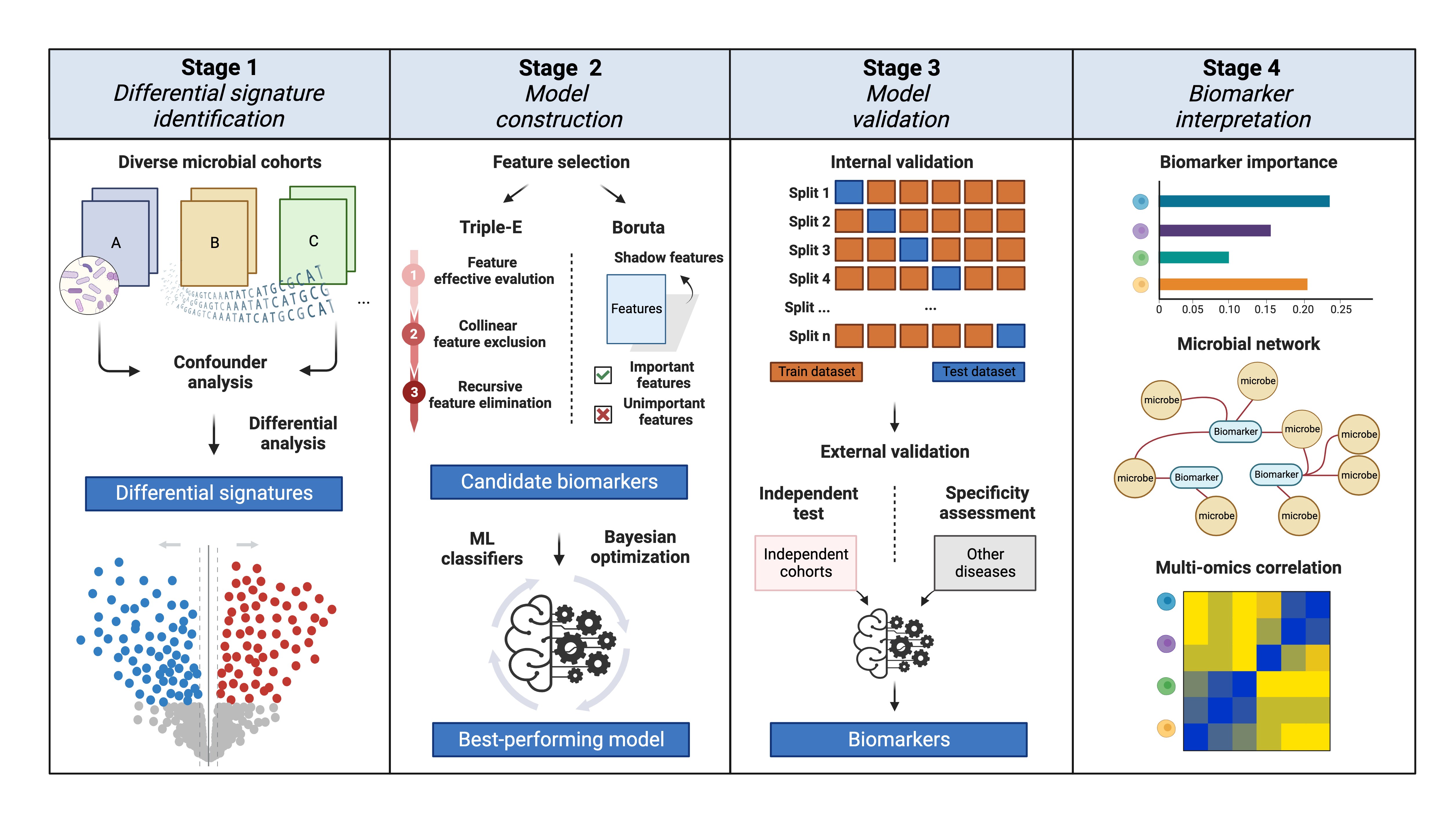
Stage1 Differential signature identification
Firstly, to mitigate challenges induced by different number of sequencing, xMarkerFinder introduces various normalization methods. In addition, rare signatures, those with low occurrence rates across cohorts are discarded to ensure that identified biomarkers are reproducible and could be applied to prospective cohorts. After data normalization and filtering, Permutational multivariate analysis of variance (PERMANOVA) test is performed. For each metadata variable, coefficient of determination (R2) value and p value are calculated to explain how variation is attributed. To identify disease or trait-associated microbial signatures across cohorts, MMUPHin is employed. Signatures with consistently significant differences in meta-analysis are identified as cross-cohort differential signatures and used for further feature selection in subsequent stages.
Stage 2 Model construction
xMarkerFinder integrates multiple widely used ML algorithms, including Logistic Regression (LR, L1 and L2 regularization), Support Vector classifier (SVC) with the Radial Basis Function kernel, K-nearest Neighbors (KNN) classifier, Decision Tree (DT) classifier, RF classifier, and Gradient Boosting (GB) classifier. All classifiers are assessed by constructing classification models with default parameters based on differential signatures.
Based on differential signatures from all cohorts in the training set and the selected ML classifier, xMarkerFinder presents the Triple-E feature selection procedure to identify an optimal panel of candidate biomarkers, including feature effectiveness evaluation, collinear feature exclusion, and recursive feature elimination. Alternatively, we also integrate the Boruta feature selection procedure. For more details, please see our Tutorial page.
Based on the selected classifier, candidate biomarkers are used to construct a classification model with stratified five-fold cross-validation to exhibit their predictive abilities, the hyperparameters of which are tuned via bayesian optimization method. The general performance of the best-performing model is reported with several metrics, including AUC, AUPR, MCC, accuracy, precision, specificity, sensitivity, and F1 score.
Stage 3 Model validation
To ensure the robustness and generalization of established candidate biomarkers and the best-performing model among geographically distinct cohorts, xMarkerFinder provides multi-perspective internal and external validations.
For internal validations, xMarkerFinder assesses candidate biomarkers’ robustness and generalizability at different level within the discovery dataset, including intra-cohort, cohort-to-cohort, and LOCO validations.
For external validations, xMarkerFinder provides an independent test and two specificity assessment tools. The independent test is conducted by applying the best-performing model to independent test datasets, which ensures their potential applicability in future prospective settings. To mitigate the false positive issues in real-world applications, xMarkerFinder integrates two specificity assessments. First, candidate biomarkers are used to construct similar classification models in other similar but non-target datasets and those with relatively worse performances in other scenarios are deemed specific (biomarker specificity assessment). Moreover, samples randomly selected from each case and control condition of non-target datasets are labelled as “control” and added into the classification model, and the variations of corresponding AUCs are calculated. The difference between the AUCs of adding external cases and the AUCs of adding external controls could be seen as a proxy of model’s specificity, and those with similar AUCs are considered specific (model specificity assessment). To enhance user convenience, we've separated the specificity assessment into a standalone module on our website. If you don't have the non-target datasets required for this analysis, you can consider this step optional. However, we still recommend conducting it, and you can find the necessary data within the public datasets we provide on the Resource page.
Stage 4 Biomarker interpretation
In addition to biomarker identification and model construction, we recommend multiple biomarker interpretation steps to better characterize microbial biomarkers’ contributions to the classification model and their potential roles in trait stratification or disease initiation and progression, including biomarker importance, microbial co-occurrence network, and multi-omics correlation. For more details, please see the Tutorial page.
For any queries or concerns related to xMarkerFinder, please contact our support team at cadd.tongji.edu.cn or webmaster@picb.ac.cn
If you find our website useful, please cite: Wenxing Gao#, Weili Lin#, Qiang Li#, Wanning Chen, Wenjing Yin, Xinyue Zhu, Sheng Gao, Lei Liu, Wenjie Li, Dingfeng Wu, Guoqing Zhang*, Ruixin Zhu*, Na Jiao*. (2024). Identification and validation of microbial biomarkers from cross-cohort datasets using xMarkerFinder, Nature Protocols 19, 2803-2830, https://doi.org/10.1038/s41596-024-00999-9.
Wu, Y., et al. (2021). "Identification of microbial markers across populations in early detection of colorectal cancer." Nat Commun 12(1): 3063. [https://doi.org/10.1038/s41467-021-23265-y]
Liu, N.-N., et al. (2022). "Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts." Nature Microbiology. 7(1): 238–250 [https://doi.org/10.1038/s41564-021-01030-7]
Gao, S., et al. (2023). "Microbial genes outperform species and SNVs as diagnostic markers for Crohn's disease on multicohort fecal metagenomes empowered by artificial intelligence." Gut Microbes 15(1): 2221428. [https://doi.org/10.1080/19490976.2023.2221428]
Gao, W., et al. (2023). "Multimodal metagenomic analysis reveals microbial single nucleotide variants as superior biomarkers for early detection of colorectal cancer." Gut Microbes 15(2): 2245562. [https://doi.org/10.1080/19490976.2023.2245562]
At xMarkerFinder, we are continuously striving to improve, fix bugs, and incorporate user feedback to make our tool more robust and user-friendly. Below you’ll find a detailed log of our updates, ensuring you’re always informed about our latest enhancements.
Latest version: 1.0.16
New features:
New visualization tools and optimized serve response time.
Enhanced Help Page with detailed instructions and examples.
Note: Always ensure to regularly back up your results and refer to our Help Page for more details.