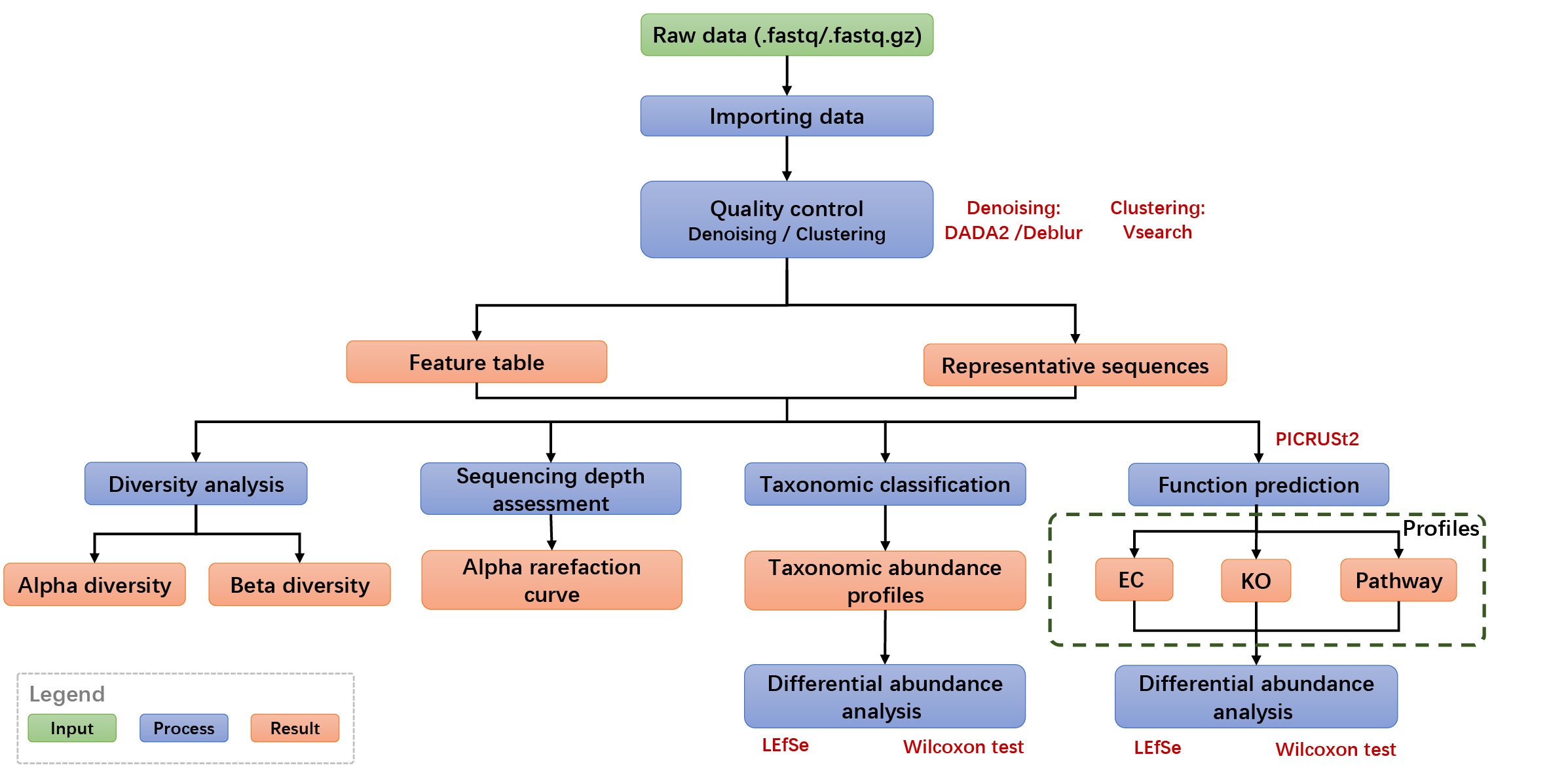
The 16S rRNA analysis pipeline, based on Qiime2, enables researchers in seamlessly completing all analyses from raw DNA sequence data to common downstream statistical results with a single click.
Species Diversity Study
Alpha diversity measures the species richness and evenness within a single sample, reflecting the complexity and diversity of the community. Beta diversity assesses the differences in species composition between samples, allowing for the comparison of similarities and dissimilarities across different communities.
Differential Flora Study
The Wilcoxon rank sum test and LEfSe are used to identify differential flora at any taxonomic level between groups.
Differential Function Study
Using representative sequences and feature abundance profiles, functional abundance profiles for Enzyme Commission numbers, KEGG ORTHOLOGY, and pathways are obtained through function prediction. Then, LEfSe is used to identify differential functions between groups.
16S rRNA Analysis Pipeline Steps (for more information, please refer to the user manual)
1、Register an account. If you don’t have an iMAC account, please click here to register your account.
2、Login. If you already have an iMAC account, please click login in the navigation bar.
3、Upload data. Download the FTP client software FileZilla, and log in sftp://fms.biosino.org:44399 using the registered user name and password. Then, upload your data through FileZilla and browse them in user center/My Data module.
4、Create new task. In the user center/Task List/16S rRNA Task, click “New Task” to create a 16S rRNA pipeline task. Then, adjust the default parameters according to your requirement and submit.
5、Download results. After the task is finished, you can download the results in the user center/Task List/16S rRNA Task and click an eye icon to view interactive graphics.