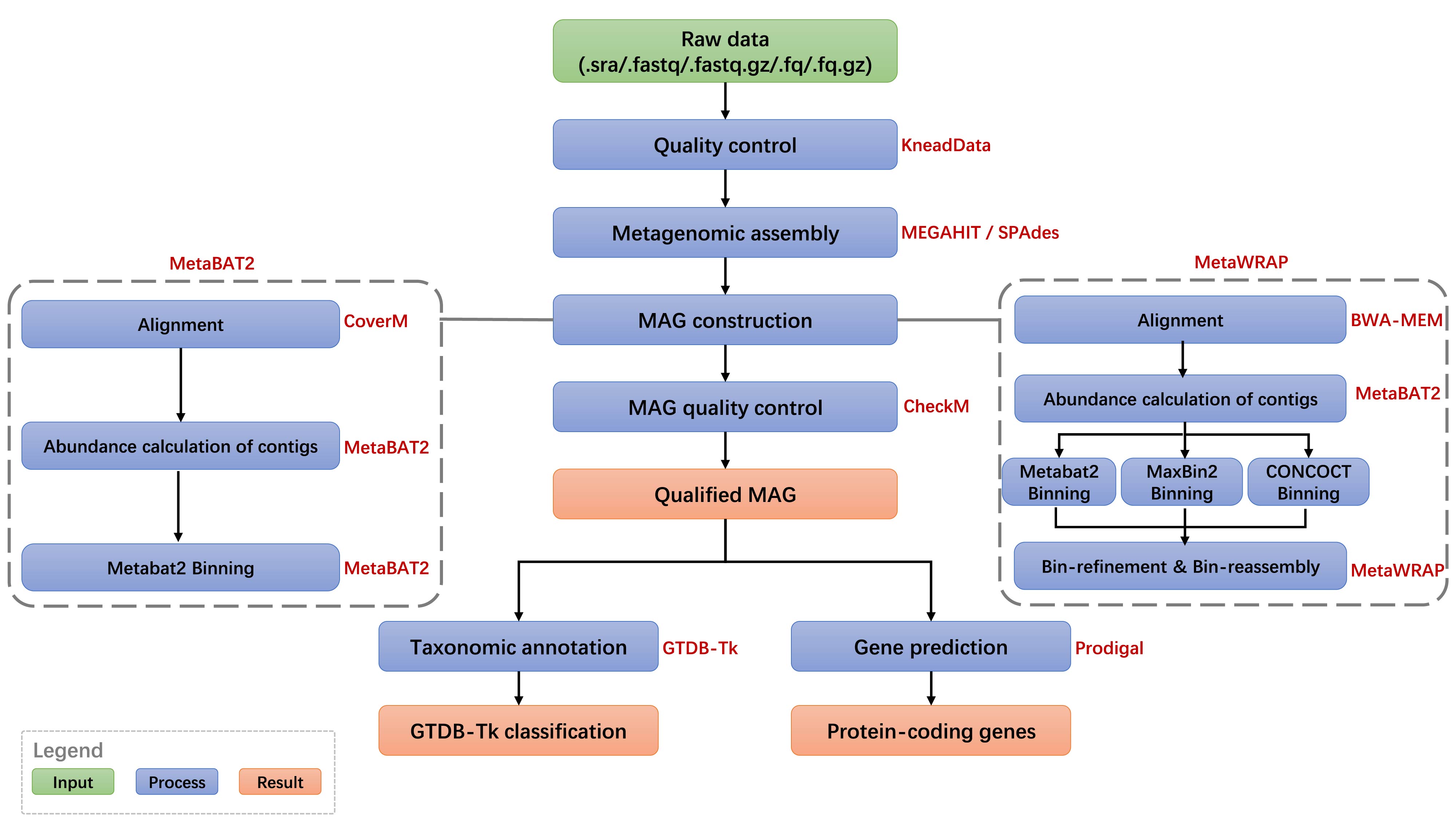
The mWGS-MAG pipeline is based on metagenome-assembled genome method. Firstly, quality control of the raw data is performed by Trimmomatic, and MEGAHIT/SPAdes is used for metagenomic assembly. Then, the MAG is reconstructed, mainly including alignment between reads and assembled contigs (CoverM), abundance calculation of contigs (MEGAHIT or SPAdes), binning (MetaBAT2 or MetaWRAP), MAG quality control (CheckM), prediction of protein-coding genes (Prodigal) and taxonomyic annotation (GTDB-Tk).
The steps of mWGS-MAG pipeline (for more information, please refer to user manual)
1、Register an account. If you don’t have an iMAC account, please click here to register your account.
2、Login. If you already have an iMAC account, please click login in the navigation bar.
3、Upload data. Download the FTP client software FileZilla, and log in sftp://fms.biosino.org:44399 using the registered user name and password. Then, upload your data through FileZilla and browse them in user center/My Data module.
4、Create new task. In the user center/Task List/mWGS-MAG Task, click “New Task” to create a mWGS-MAG pipeline task. Then, adjust the default parameters according to your requirement and submit.
5、Download results. After the task is finished, you can download the results in the user center/Task List/mWGS-MAG Task and click the eye icon to view interactive graphics.