1. Catalytic bioparts
Catalytic bioparts are fundamental to the design, construction, and optimization of biological systems for specific metabolic pathways. Catalytic bioparts not only refer to enzymes or other biomolecules that can catalyze biochemical reactions, but also their performance data and nuclear acid sequences in specific chassis( [1],[2] ). The selection and utilization of catalytic bioparts directly influence the efficiency and specificity of biosynthetic systems([3]). By precise designing and assembling various catalytic bioparts, novel biological systems can be created for the production of valuable chemicals, pharmaceuticals, biofuels, and other products([4],[5],[6])
2. Motivation of RDBSB database
Constructing biosynthetic systems needs to assemble target biosynthetic pathways within specific chassis in a “plug-and-play” manner based on standardized catalytic bioparts([7],[8]) . Thus, a reliable database of experimentally validated catalytic bioparts is essential for providing the resources required to design and optimize these biosynthetic systems. The Registry of Standard Biological Parts includes over 300 catalytic bioparts developed to address specific challenges in iGEM competition([9]). While databases like Swiss-Prot, MetaCyc, KEGG, BRENDA, CAZy, and GTDB, focus on curated protein ( [10] ), biosynthetic pathways ( [11],[12] ), or enzyme ( [13],[14],[15] ) in non-engineered organism, they lack crucial information on the performance of catalytic biopart within chassis, which is vital for synthetic biology applications.
3. Information integrity levels
The integrity of catalytic biopart information is classified into four levels:
Level 1 refers to catalytic bioparts that contain protein or coding sequences.
Level 2 includes bioparts from Level 1 that have associated reactions.
Level 3 comprises bioparts from Level 2 with experimentally validated reactions.
Level 4 consists of bioparts from Level 3 that also have optimum pH, optimum temperature, and chassis information.
4. Data Content of RDBSB database
138 data elements for catalytic biopart were established by extracting the bio-chemical reactions and kinetic parameters related data element from BRENDA, Swiss-Prot, MetaCyc, KEGG, RHEA, and BMKS, and combining the data elements used in the lab of synthetic biology. Data elements of protein, enzyme, compound, and reaction were adapted to according data elements of Uniprot, Enzyme, ChEBI, and Rhea, respectively.
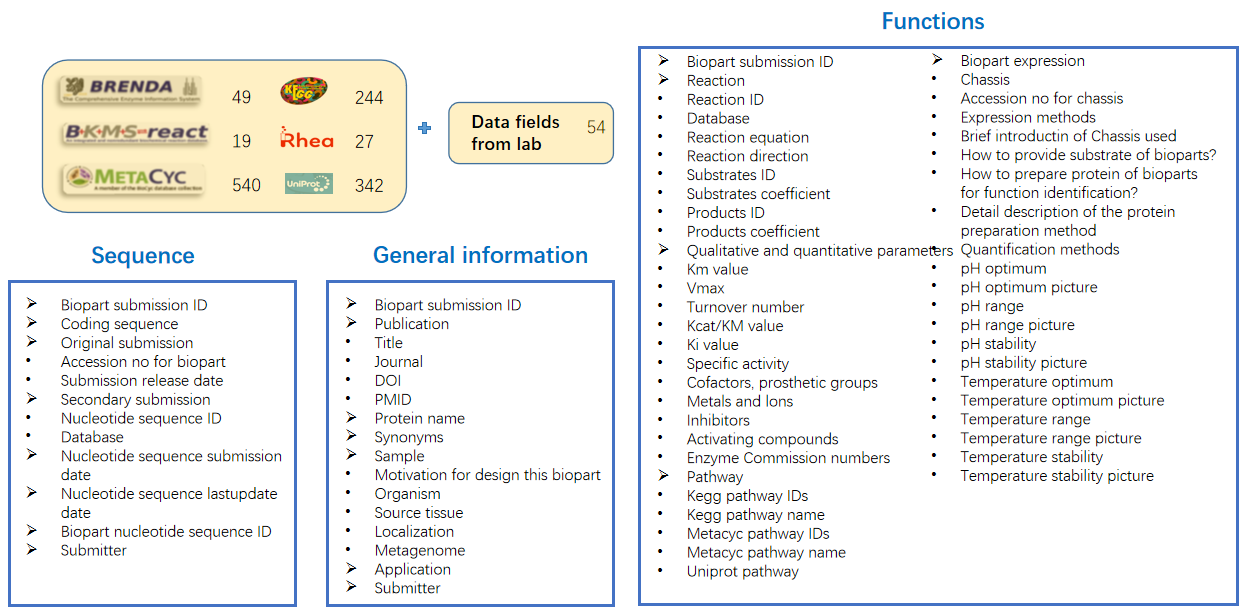
5. Data sources
MetaCyc Protein, BRENDA, Swiss-Prot, KEGG GENES and NCBI GenBank served as the primary resources for integrating catalytic bioparts, providing crucial information on proteins, coding sequences (CDS), reactions, and functions ( [10],[11],[12],[14],[16] ). Reactions were sourced from MetaCyc Reactions, MetaCyc Enzrxns, KEGG REACTION, RHEA, and BKMS ( [17],[18] ), while substrate and product annotations were derived from MetaCyc Compounds, KEGG COMPOUND, PubChem, and ChEBI ( [19],[20] ). KEGG Enzyme, ENZYME, Pfam, MetaCyc pathways, KEGG PATHWAY, AlphaFoldDB, and NCBI Taxonomy were used to annotate the enzyme, domain, pathway, structure, and organism associated with each biopart ( [21],[22],[23],[24] ). The literatures curated by RDBSB were sourced from PubMed ([25]).
6. Usage of RDBSB
6.1 Keywords Search
Search by catalytic biopart name: e.g. salutaridine synthase
Search by catalytic biopart ID: e.g. OENC102406
Search by IDs from other data integrated databases: e.g. Q842E2, P9WN65, CHEBI:57540
Search by compound: e.g. (2E)-geranyl diphosphate, palmitoyl-CoA
Search by Pathway: e.g. Carbohydrate biosynthesis
Search by Chassis: e.g Escherichia coli
6.2 Sequence based search
Search by amino acid sequence6.3 Graph database based search
Search by MapView6.4 Browse and filter
The browse and filter page lists all potentially catalytic bioparts based on the user's query. Users can use the information integrity filtering, evidence filtering, and sorting tools (optimum pH, optimum temperature, chassis name, gene name, organism, EC number, Pathway name, and compound) at the right side of the page to find catalytic biopart of interest.
Here, we filter the catalytic biopart with optimum pH range of 4.0-9.0, optimum temperature range of 20 ºC -40 ºC, chassis of Escherichia coli, reactions with experimental evidence, in total, we get 2097 catalytic bioparts.
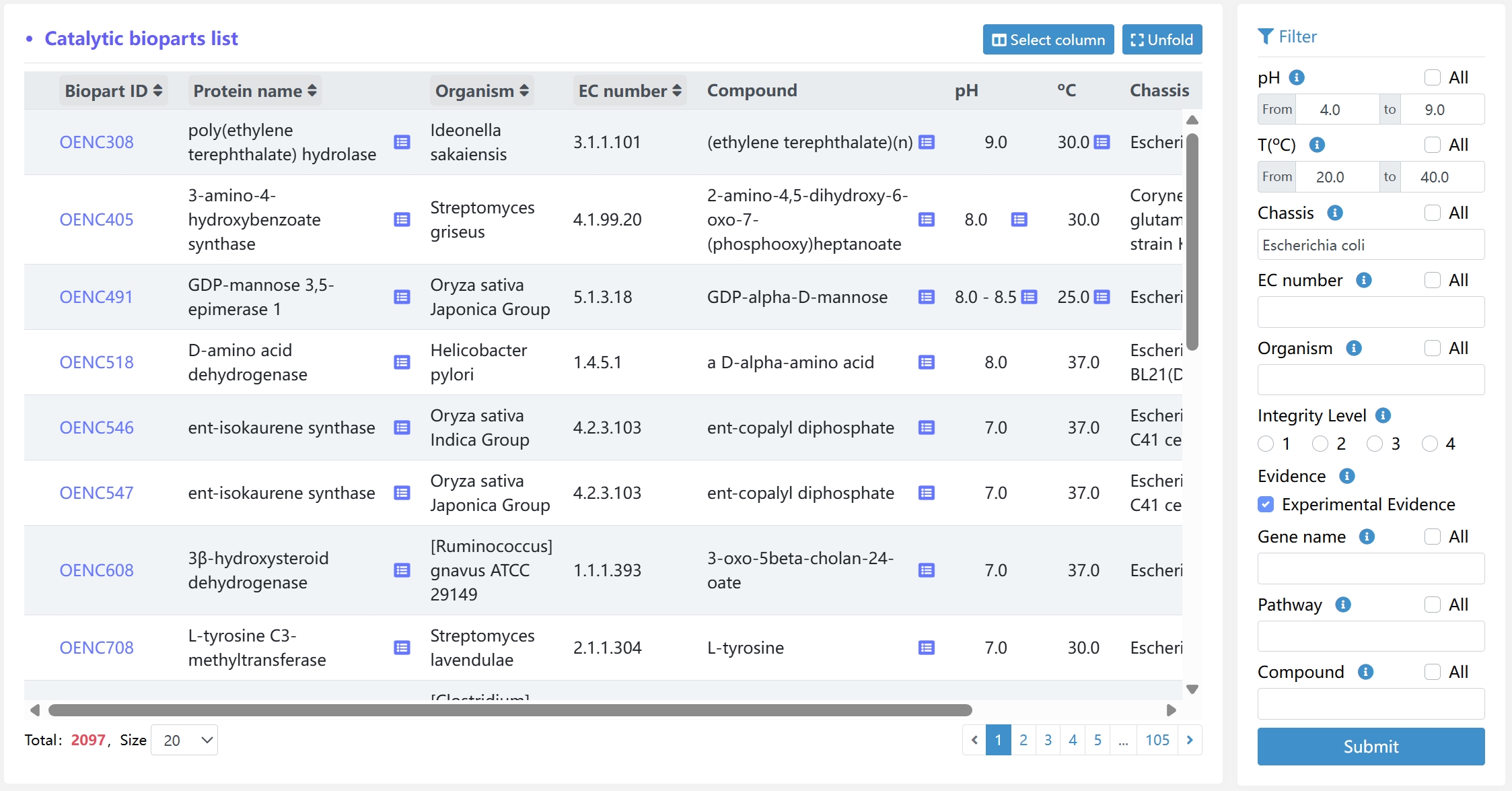
6.5 Detailed View
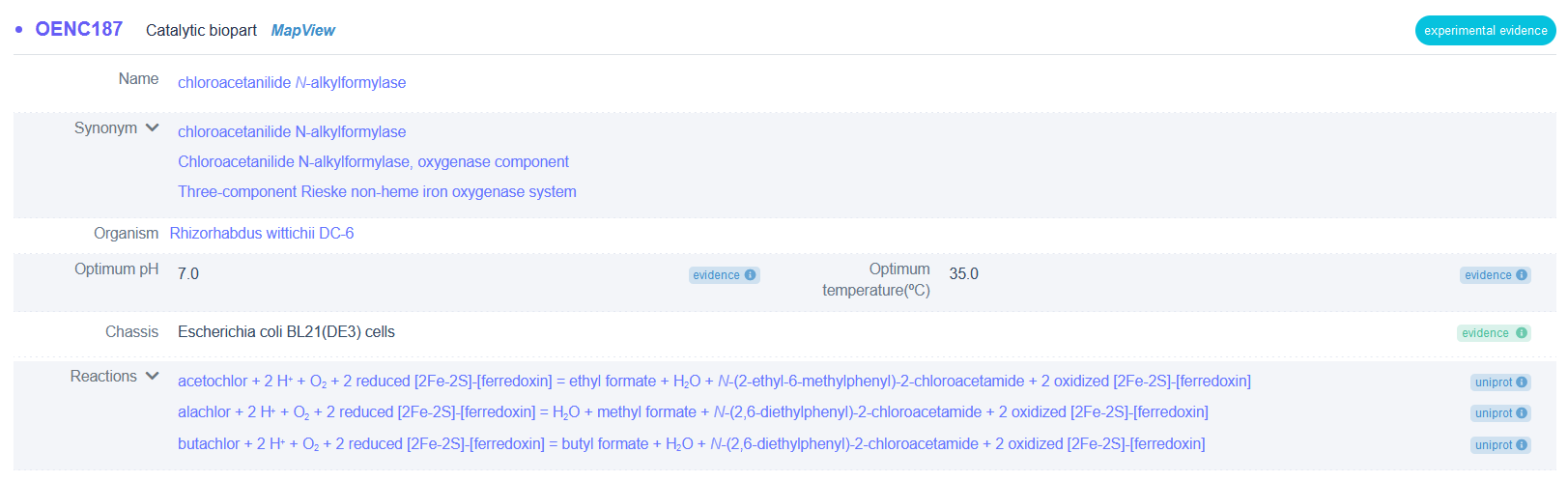
View catalytic biopart detailed information, such as basic information, function information (including catalytic functions, qualitative and quantitative parameters, and biopart expression), sequence information (nucleotide sequence and amino acid sequence) and more. We provided visualization of multidimensional information of catalytic bioparts Mol* Viewer ( [26] ).
(1) Example of basic information of chloroacetanilide N-alkylformylase (biopart ID: OENC187)
(2) Example of catalytic functions of ent-isokaurene synthase (biopart ID: OENC547)
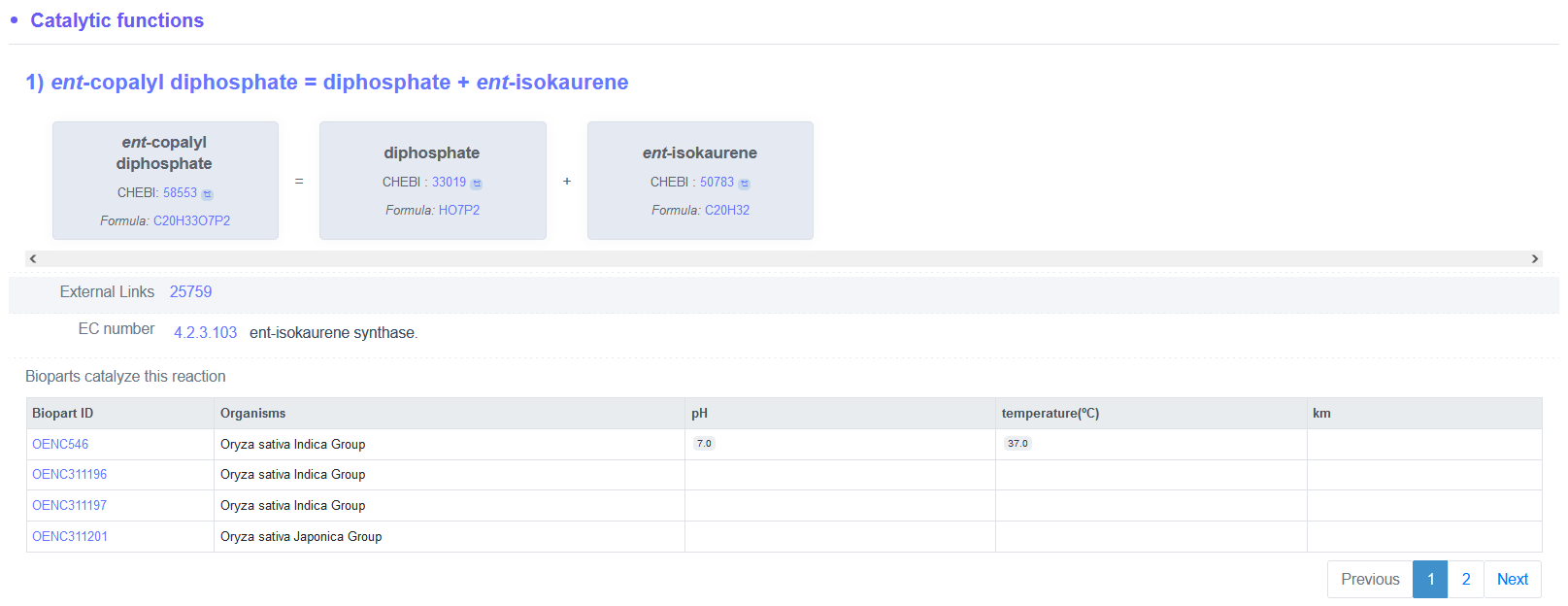
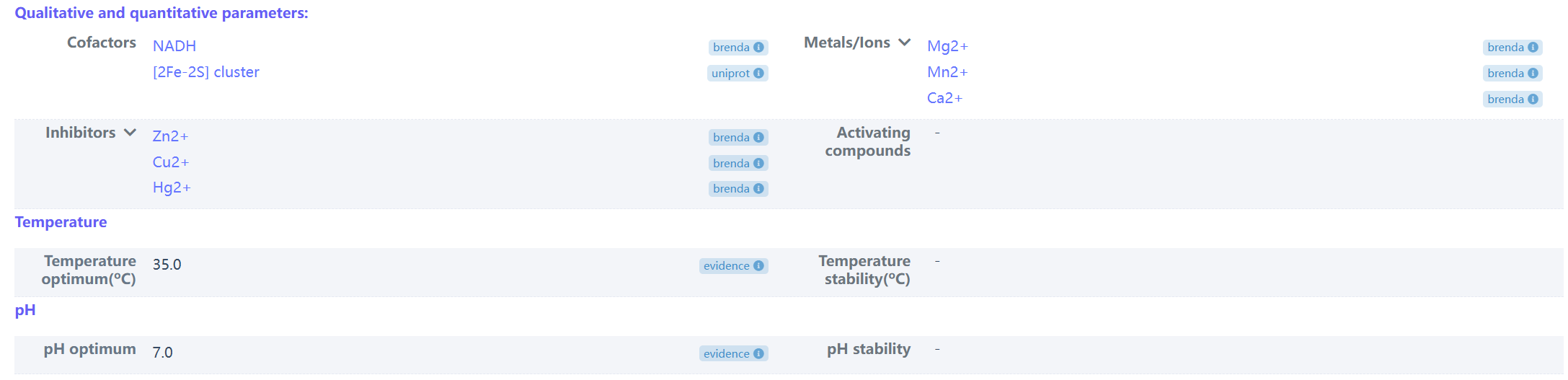
(3) Example of qualitative and quantitative parameters of chloroacetanilide N-alkylformylase (biopart ID: OENC187)
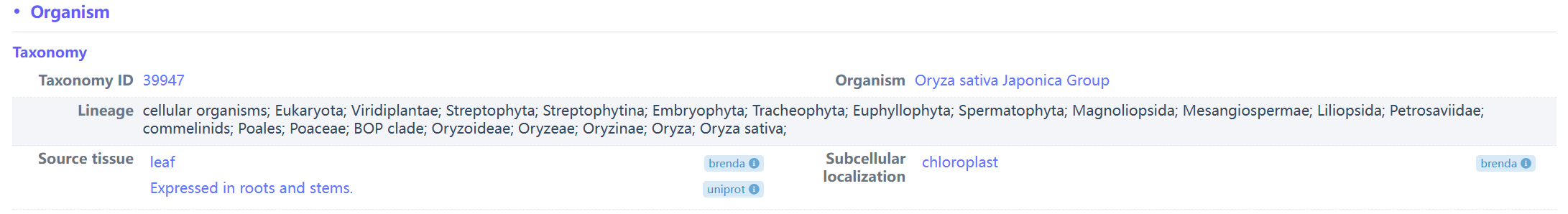
(4) Example of source organism of ent-isokaurene synthase (biopart ID: OENC547)
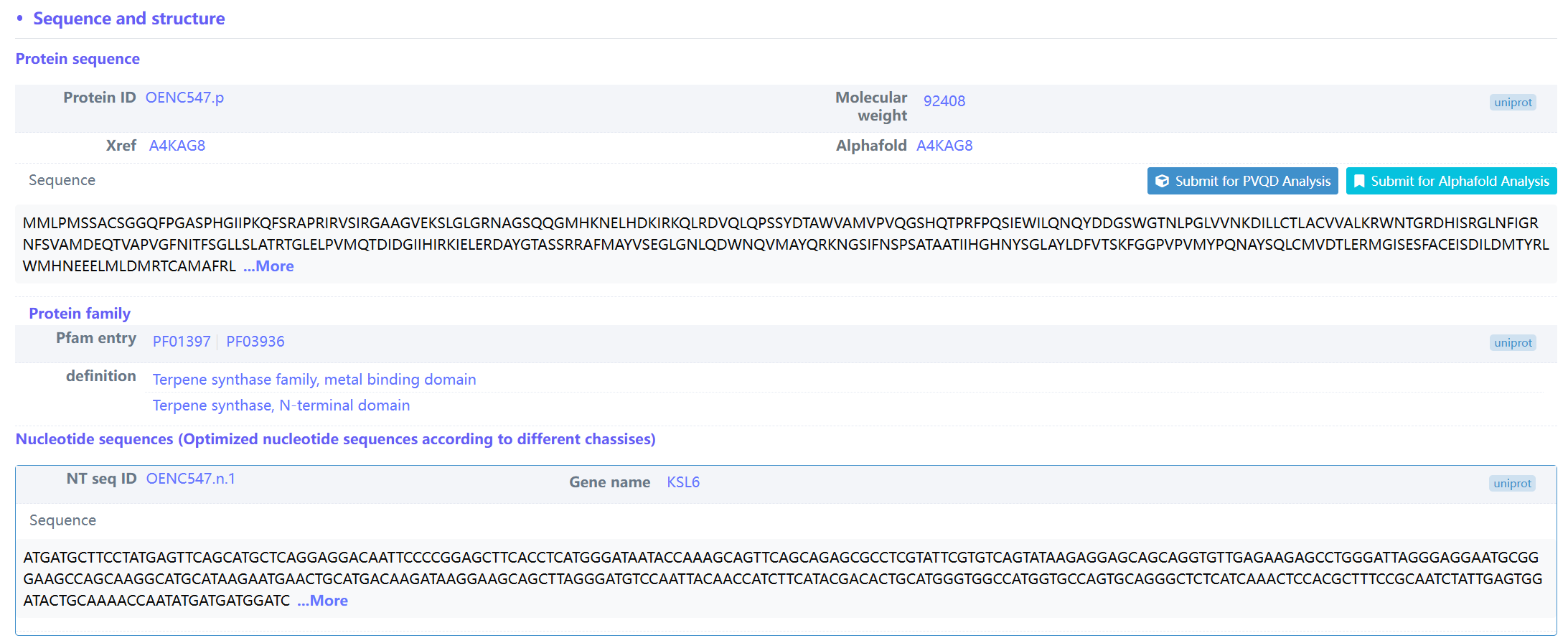
(5) Example of sequence and structure of ent-isokaurene synthase (biopart ID: OENC547)
6.6 Structure prediction
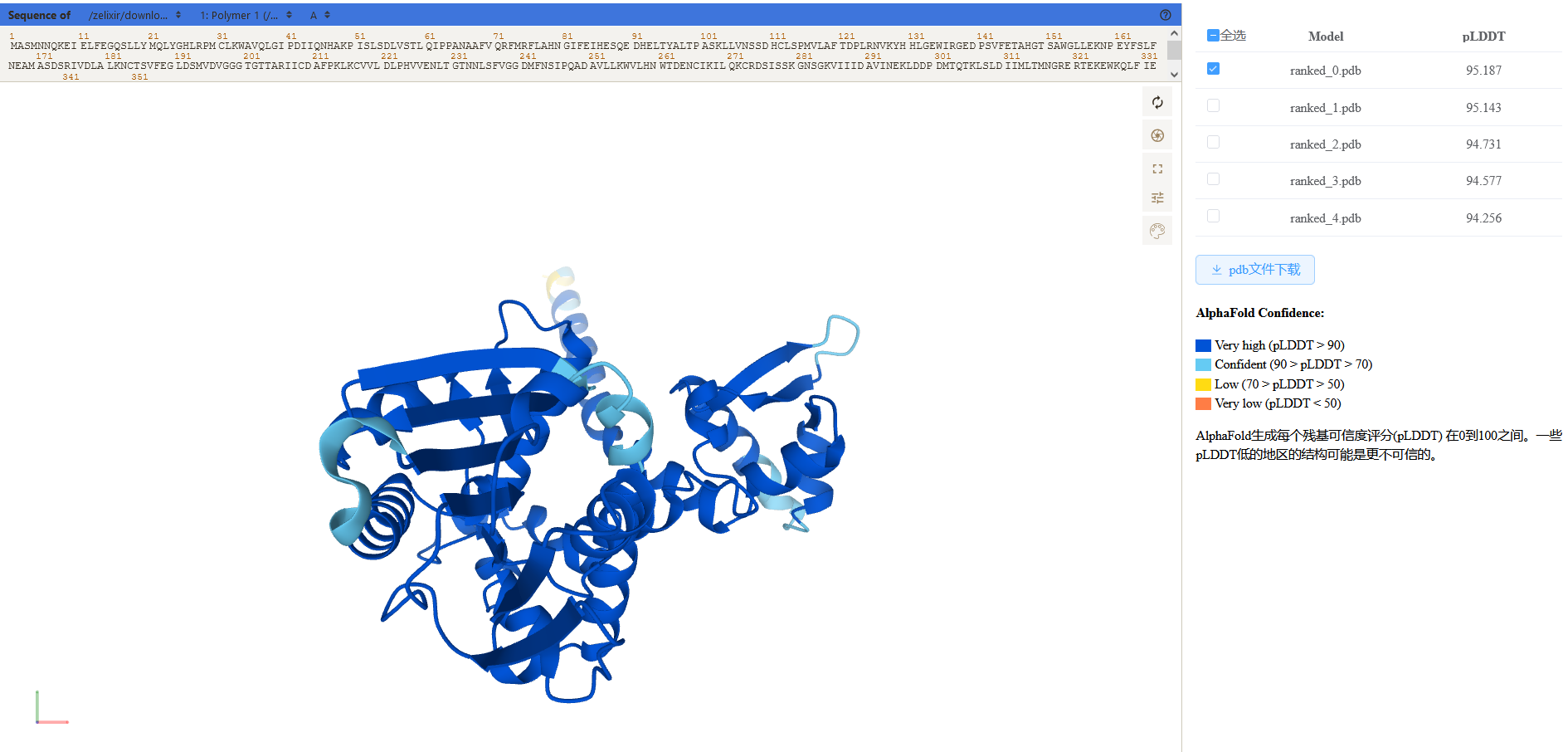
(1) Example of structure prediction by AlphaFold, eg. GmOMT2 (biopart ID: OENC19)
By click the “Submit for AlphaFold Analysis” above the amino acid sequence of biopart in the detail page, the user could enter the email address to receive the analysis result and the amino acid sequence of GmOMT2 will enter the structure prediction analysis.
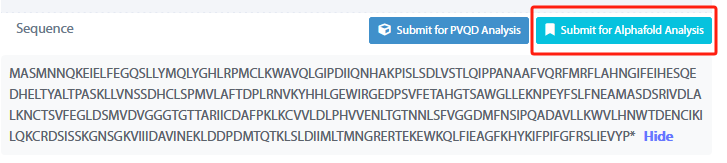
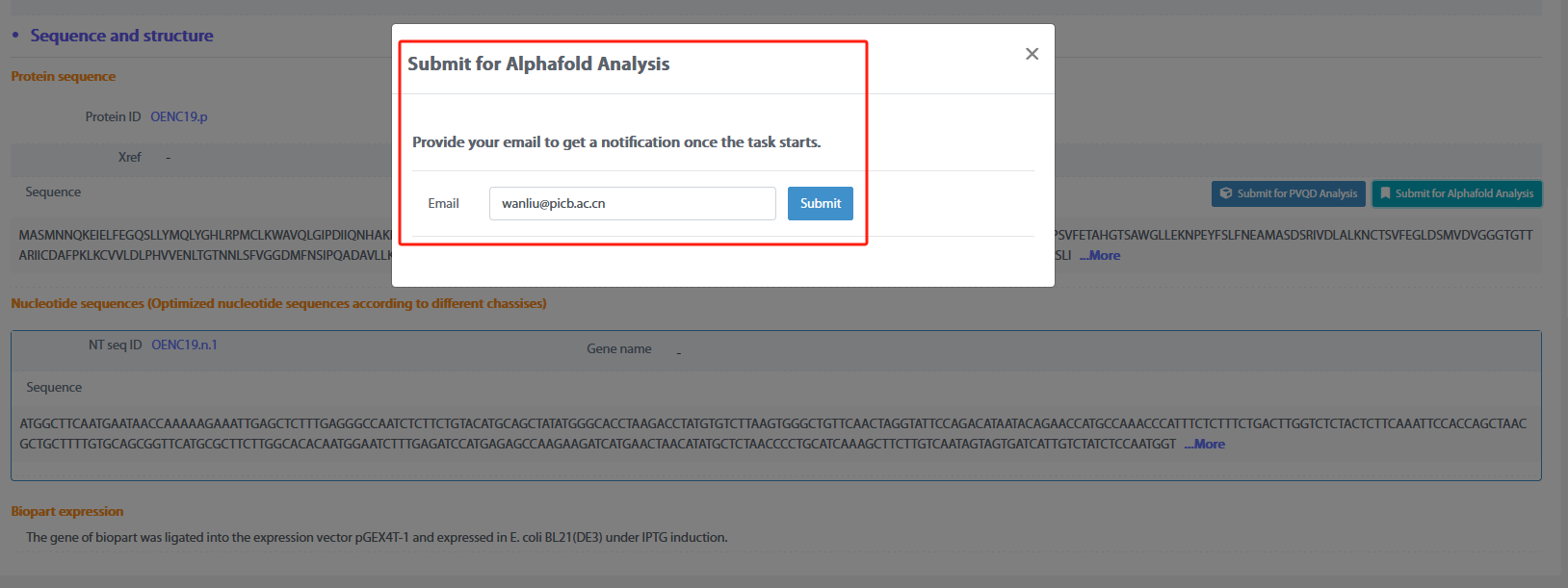
After the analysis is done, the user will receive a link by email for the visualization of the 3D structure.
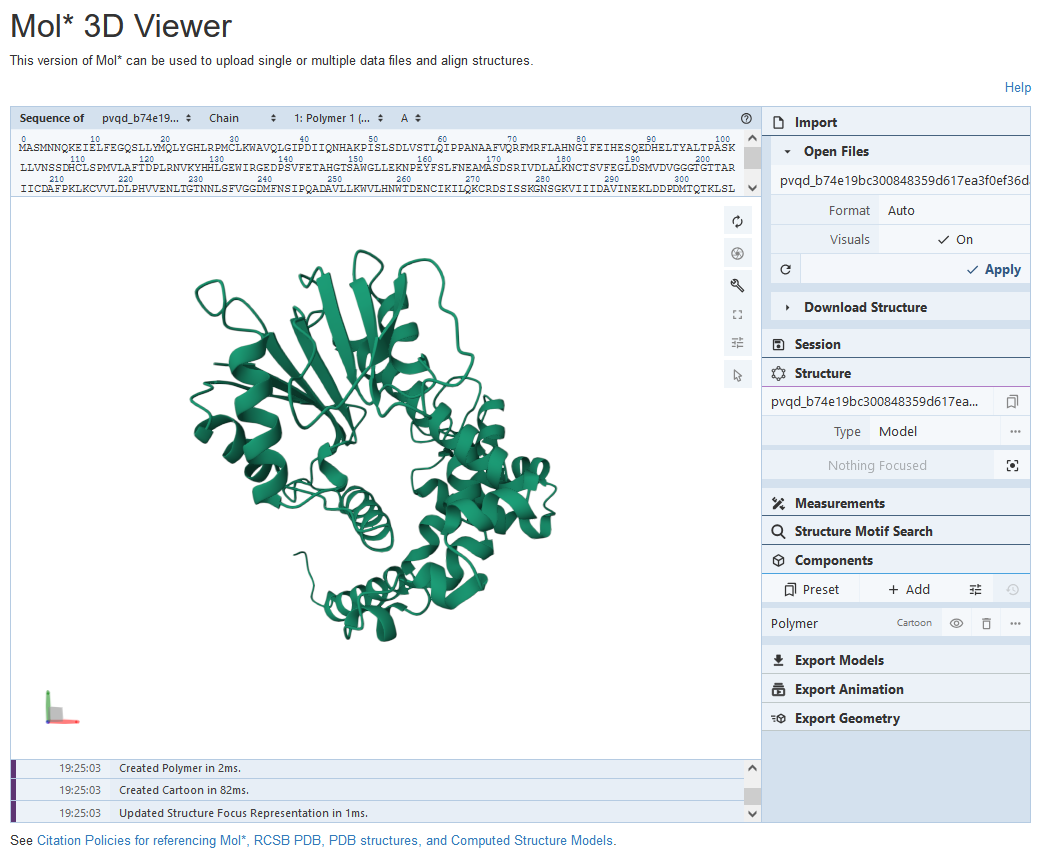
(2) Example of structure prediction by PVQD, eg. GmOMT2 (biopart ID: OENC19)
By click the “Submit for PVQD Analysis” above the amino acid sequence of biopart in the detail page, the user could enter the email address to receive the analysis result and the amino acid sequence of GmOMT2 will enter the structure prediction analysis.
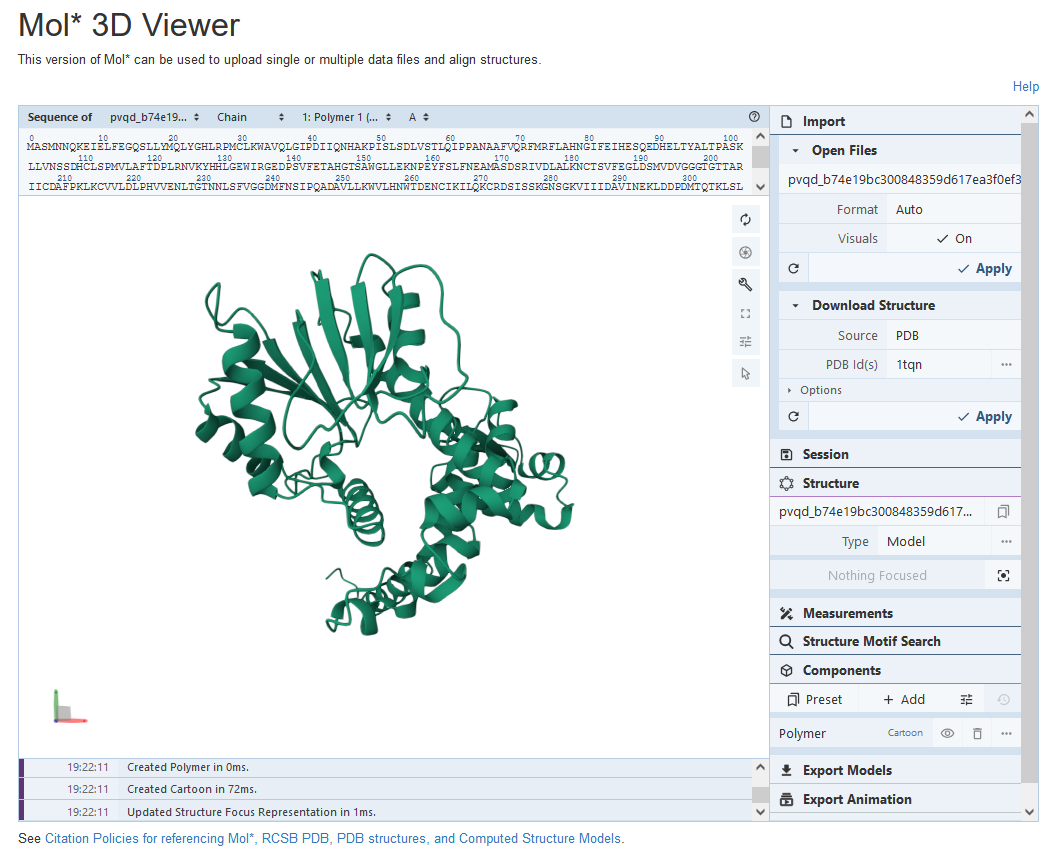
After the analysis is done, the user will receive a link by email for the visualization of the two different conformations of GmOMT2, and the structure could be view by the public structure tool (25).
The first conformation of GmOMT2 is shown as below:
The second conformation of GmOMT2 is displayed as below:
6.7 PathFinder
PathFinder is available in the home page of RDBSB under the Tools category.
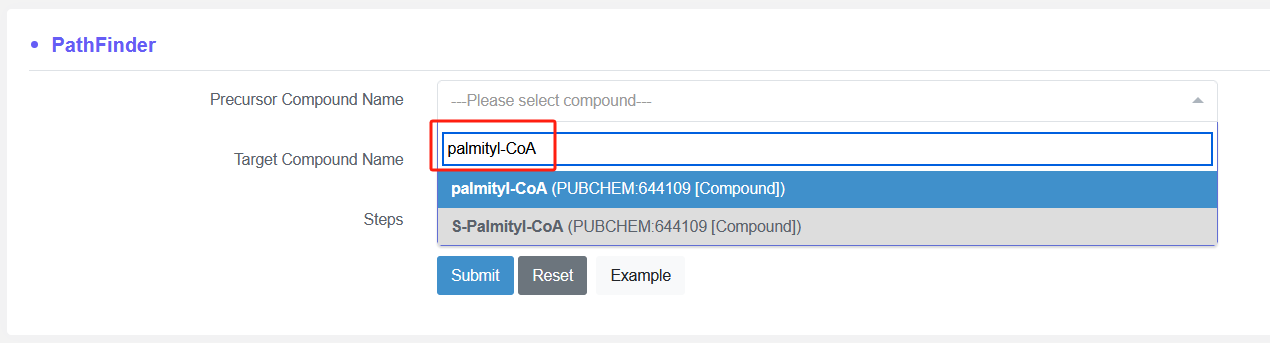
By enter the precursor compound name and target compoud name, the possible pathway will be design by this tool.
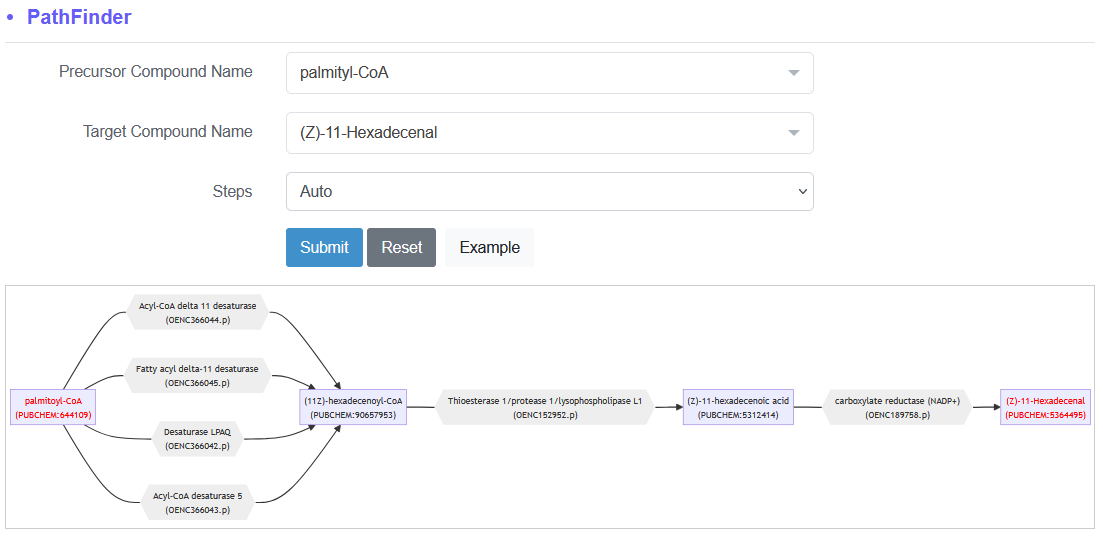
Below is an example of four possible pathway design by Path Finder after inputting palmitoyl-CoA and (Z)-11-Hexadecenal as the precursor compound and target compoud, respectively.


 Related
Related